You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001540_00421
You are here: Home > Sequence: MGYG000001540_00421
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Corynebacterium provencense | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Mycobacteriales; Mycobacteriaceae; Corynebacterium; Corynebacterium provencense | |||||||||||
| CAZyme ID | MGYG000001540_00421 | |||||||||||
| CAZy Family | GT39 | |||||||||||
| CAZyme Description | putative dolichyl-phosphate-mannose--protein mannosyltransferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 461633; End: 463240 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT39 | 40 | 301 | 4.4e-64 | 0.9910313901345291 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1928 | PMT1 | 1.84e-40 | 27 | 534 | 33 | 693 | Dolichyl-phosphate-mannose--protein O-mannosyl transferase [Posttranslational modification, protein turnover, chaperones]. |
| COG4346 | COG4346 | 1.70e-17 | 98 | 466 | 136 | 415 | Predicted membrane-bound dolichyl-phosphate-mannose-protein mannosyltransferase [Posttranslational modification, protein turnover, chaperones]. |
| pfam02366 | PMT | 1.77e-14 | 39 | 295 | 2 | 235 | Dolichyl-phosphate-mannose-protein mannosyltransferase. This is a family of Dolichyl-phosphate-mannose-protein mannosyltransferase proteins EC:2.4.1.109. These proteins are responsible for O-linked glycosylation of proteins, they catalyze the reaction:- Dolichyl phosphate D-mannose + protein <=> dolichyl phosphate + O-D-mannosyl-protein. Also in this family is Drosophila rotated abdomen protein which is a putative mannosyltransferase. This family appears to be distantly related to pfam02516 (A Bateman pers. obs.). This family also contains sequences from ArnTs (4-amino-4-deoxy-L-arabinose lipid A transferase). They catalyze the addition of 4-amino-4-deoxy-l-arabinose (l-Ara4N) to the lipid A moiety of the lipopolysaccharide. This is a critical modification enabling bacteria (e.g. Escherichia coli and Salmonella typhimurium) to resist killing by antimicrobial peptides such as polymyxins. Members such as undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase are predicted to have 12 trans-membrane regions. The N-terminal portion of these proteins is hypothesized to have a conserved glycosylation activity which is shared between distantly related oligosaccharyltransferases ArnT and PglB families. |
| pfam16192 | PMT_4TMC | 7.19e-13 | 333 | 532 | 2 | 198 | C-terminal four TMM region of protein-O-mannosyltransferase. PMT_4TMC is the C-terminal four membrane-pass region of protein-O-mannosyltransferases and similar enzymes. |
| COG1807 | ArnT | 1.46e-08 | 64 | 484 | 34 | 419 | 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AWT25677.1 | 0.0 | 1 | 535 | 1 | 535 |
| AEK37413.1 | 0.0 | 31 | 535 | 13 | 514 |
| AGP31294.1 | 0.0 | 31 | 535 | 17 | 521 |
| AHW63433.1 | 2.61e-271 | 32 | 535 | 24 | 529 |
| QQC48610.1 | 7.83e-240 | 36 | 534 | 1 | 519 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6P25_A | 1.21e-06 | 34 | 258 | 50 | 253 | Structureof S. cerevisiae protein O-mannosyltransferase Pmt1-Pmt2 complex bound to the sugar donor and a peptide acceptor [Saccharomyces cerevisiae W303],6P2R_A Structure of S. cerevisiae protein O-mannosyltransferase Pmt1-Pmt2 complex bound to the sugar donor [Saccharomyces cerevisiae W303] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8NRZ6 | 1.29e-162 | 14 | 534 | 19 | 519 | Probable dolichyl-phosphate-mannose--protein mannosyltransferase OS=Corynebacterium glutamicum (strain ATCC 13032 / DSM 20300 / BCRC 11384 / JCM 1318 / LMG 3730 / NCIMB 10025) OX=196627 GN=pmt PE=3 SV=1 |
| P9WN04 | 7.65e-135 | 35 | 534 | 45 | 521 | Probable dolichyl-phosphate-mannose--protein mannosyltransferase OS=Mycobacterium tuberculosis (strain CDC 1551 / Oshkosh) OX=83331 GN=pmt PE=3 SV=2 |
| P9WN05 | 7.65e-135 | 35 | 534 | 45 | 521 | Probable dolichyl-phosphate-mannose--protein mannosyltransferase OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=pmt PE=1 SV=2 |
| L8F4Z2 | 1.01e-133 | 39 | 534 | 43 | 515 | Probable dolichyl-phosphate-mannose--protein mannosyltransferase OS=Mycolicibacterium smegmatis (strain MKD8) OX=1214915 GN=pmt PE=3 SV=1 |
| P33775 | 6.62e-06 | 34 | 258 | 50 | 253 | Dolichyl-phosphate-mannose--protein mannosyltransferase 1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=PMT1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999829 | 0.000187 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
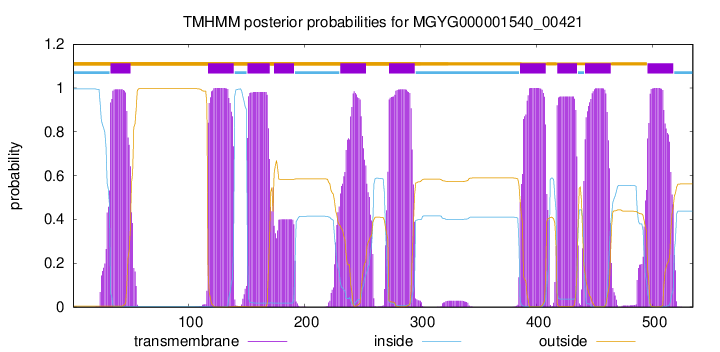
| start | end |
|---|---|
| 33 | 50 |
| 117 | 139 |
| 151 | 170 |
| 174 | 191 |
| 231 | 253 |
| 273 | 295 |
| 386 | 408 |
| 418 | 435 |
| 442 | 464 |
| 496 | 518 |
