You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001540_00595
You are here: Home > Sequence: MGYG000001540_00595
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
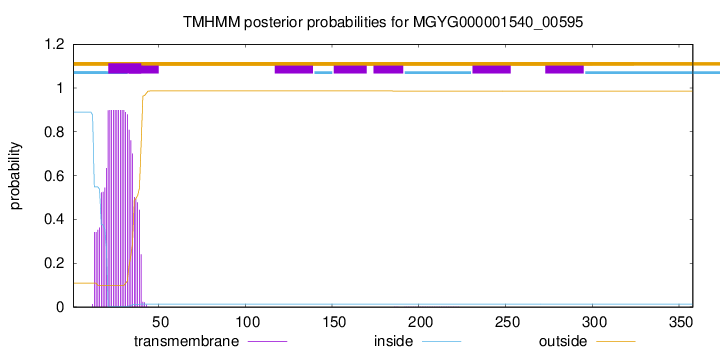
TMHMM annotations
Basic Information help
| Species | Corynebacterium provencense | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Mycobacteriales; Mycobacteriaceae; Corynebacterium; Corynebacterium provencense | |||||||||||
| CAZyme ID | MGYG000001540_00595 | |||||||||||
| CAZy Family | CE1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 660504; End: 661580 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE1 | 92 | 257 | 6.7e-26 | 0.6079295154185022 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3509 | LpqC | 3.86e-36 | 86 | 358 | 38 | 311 | Poly(3-hydroxybutyrate) depolymerase [Secondary metabolites biosynthesis, transport and catabolism]. |
| COG4099 | COG4099 | 2.84e-09 | 99 | 244 | 178 | 329 | Predicted peptidase [General function prediction only]. |
| COG0400 | YpfH | 1.42e-07 | 107 | 266 | 13 | 182 | Predicted esterase [General function prediction only]. |
| COG1506 | DAP2 | 1.74e-07 | 19 | 218 | 313 | 508 | Dipeptidyl aminopeptidase/acylaminoacyl peptidase [Amino acid transport and metabolism]. |
| COG0412 | DLH | 4.92e-04 | 82 | 244 | 1 | 172 | Dienelactone hydrolase [Secondary metabolites biosynthesis, transport and catabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ALO15718.1 | 3.19e-25 | 84 | 358 | 20 | 293 |
| SBO97871.1 | 7.51e-20 | 83 | 284 | 65 | 284 |
| ASY31590.1 | 9.36e-20 | 79 | 358 | 173 | 455 |
| ATB27233.1 | 8.85e-19 | 92 | 354 | 219 | 485 |
| QZZ25148.1 | 1.13e-17 | 77 | 309 | 44 | 270 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9Y871 | 4.86e-14 | 95 | 309 | 292 | 518 | Feruloyl esterase B OS=Piromyces equi OX=99929 GN=ESTA PE=2 SV=1 |
| A2QYU7 | 1.85e-10 | 138 | 309 | 95 | 247 | Probable feruloyl esterase C OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=faeC PE=3 SV=1 |
| A1CC33 | 2.05e-09 | 161 | 305 | 114 | 257 | Probable feruloyl esterase C OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=faeC-2 PE=3 SV=1 |
| Q7RWX8 | 7.71e-09 | 63 | 300 | 25 | 256 | Feruloyl esterase D OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=faeD-3.544 PE=1 SV=1 |
| B8N7Z6 | 1.19e-08 | 95 | 306 | 43 | 256 | Probable feruloyl esterase C OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=faeC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.165204 | 0.801769 | 0.020763 | 0.010606 | 0.001152 | 0.000486 |