You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001540_01177
You are here: Home > Sequence: MGYG000001540_01177
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Corynebacterium provencense | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Mycobacteriales; Mycobacteriaceae; Corynebacterium; Corynebacterium provencense | |||||||||||
| CAZyme ID | MGYG000001540_01177 | |||||||||||
| CAZy Family | GT83 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1259808; End: 1261973 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT83 | 89 | 490 | 3.7e-34 | 0.6648148148148149 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13231 | PMT_2 | 4.23e-23 | 90 | 248 | 1 | 159 | Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
| COG1807 | ArnT | 1.62e-16 | 73 | 521 | 46 | 445 | 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| pfam09594 | GT87 | 0.001 | 92 | 262 | 12 | 154 | Glycosyltransferase family 87. The enzymes in this family are glycosyltransferases. PimE is involved in phosphatidylinositol mannoside (PIM) synthesis, a major class of glycolipids in all mycobacteria. PimE is a polyprenol-phosphate-mannose-dependent mannosyltransferase that transfers the fifth mannose of PIM. The family also includes alpha(1-->3) arabinofuranosyltransferase, invloved in the synthesis of of mycobacterial arabinogalactan. |
| COG4745 | COG4745 | 0.002 | 93 | 205 | 67 | 178 | Predicted membrane-bound mannosyltransferase [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QWZ09319.1 | 5.19e-102 | 34 | 386 | 38 | 380 |
| QUB37647.1 | 1.04e-98 | 28 | 720 | 198 | 796 |
| QHU89858.1 | 2.20e-79 | 37 | 717 | 275 | 902 |
| QHU92512.1 | 5.11e-77 | 37 | 717 | 273 | 899 |
| QHU90800.1 | 7.06e-77 | 37 | 717 | 275 | 899 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O34575 | 6.96e-59 | 37 | 702 | 12 | 660 | Putative mannosyltransferase YkcB OS=Bacillus subtilis (strain 168) OX=224308 GN=ykcB PE=3 SV=2 |
| P37483 | 7.09e-54 | 30 | 702 | 3 | 647 | Putative mannosyltransferase YycA OS=Bacillus subtilis (strain 168) OX=224308 GN=yycA PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
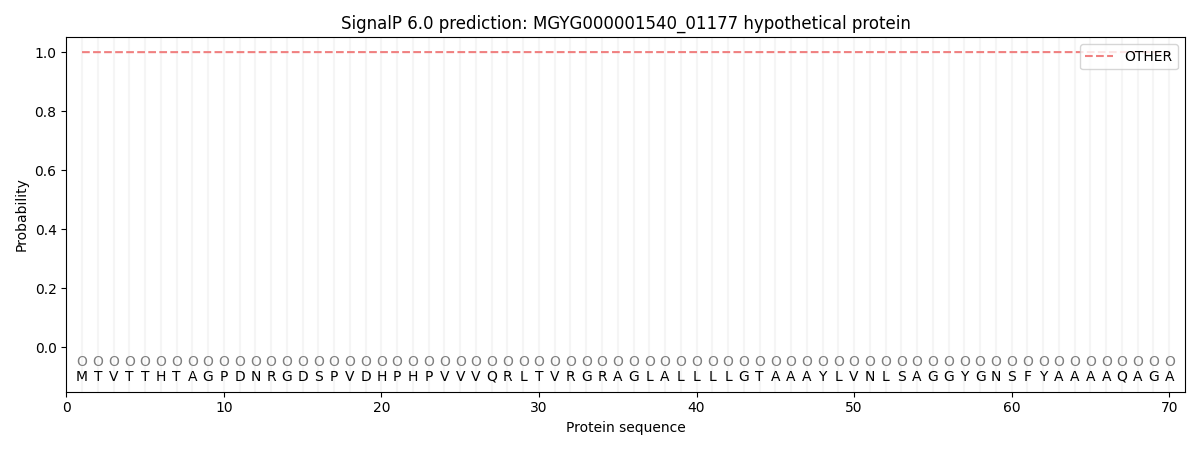
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999947 | 0.000055 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
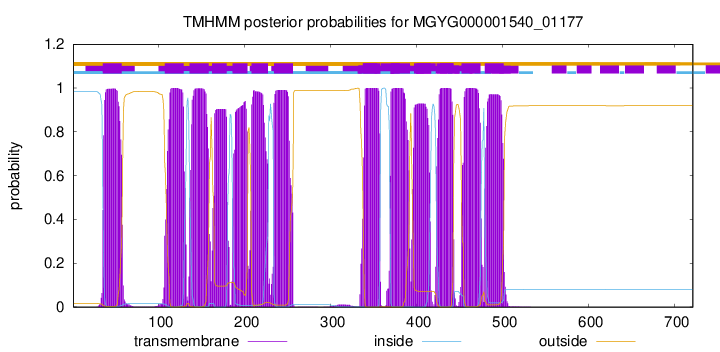
| start | end |
|---|---|
| 35 | 57 |
| 107 | 129 |
| 136 | 158 |
| 162 | 179 |
| 186 | 203 |
| 208 | 227 |
| 234 | 256 |
| 338 | 357 |
| 370 | 392 |
| 397 | 416 |
| 423 | 442 |
| 452 | 474 |
| 479 | 501 |
