You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001542_00434
You are here: Home > Sequence: MGYG000001542_00434
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
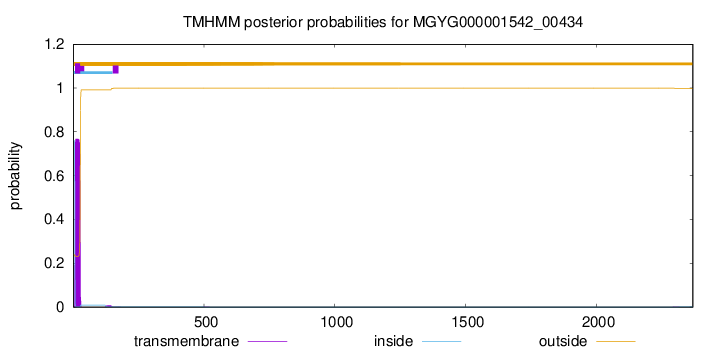
TMHMM annotations
Basic Information help
| Species | Paenibacillus_A sp900069005 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus_A; Paenibacillus_A sp900069005 | |||||||||||
| CAZyme ID | MGYG000001542_00434 | |||||||||||
| CAZy Family | CE15 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 88492; End: 95604 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 1331 | 1668 | 1.4e-94 | 0.9900990099009901 |
| CE15 | 260 | 607 | 1.4e-61 | 0.9814126394052045 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00331 | Glyco_hydro_10 | 1.33e-92 | 1331 | 1668 | 1 | 310 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 6.46e-86 | 1372 | 1666 | 1 | 263 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 1.24e-72 | 1331 | 1668 | 26 | 339 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| cd00063 | FN3 | 4.83e-14 | 1873 | 1961 | 1 | 93 | Fibronectin type 3 domain; One of three types of internal repeats found in the plasma protein fibronectin. Its tenth fibronectin type III repeat contains an RGD cell recognition sequence in a flexible loop between 2 strands. Approximately 2% of all animal proteins contain the FN3 repeat; including extracellular and intracellular proteins, membrane spanning cytokine receptors, growth hormone receptors, tyrosine phosphatase receptors, and adhesion molecules. FN3-like domains are also found in bacterial glycosyl hydrolases. |
| NF033190 | inl_like_NEAT_1 | 4.30e-12 | 2098 | 2368 | 474 | 754 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ABX44209.1 | 0.0 | 38 | 982 | 1283 | 2228 |
| QAY65909.1 | 3.51e-201 | 1536 | 2370 | 1 | 825 |
| AEV67023.1 | 1.07e-156 | 960 | 2366 | 165 | 1456 |
| QHW33615.1 | 9.76e-147 | 1326 | 2365 | 351 | 1330 |
| QAY65135.1 | 2.23e-146 | 239 | 976 | 44 | 830 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3RDK_A | 1.45e-75 | 1331 | 1669 | 7 | 338 | Proteincrystal structure of xylanase A1 of Paenibacillus sp. JDR-2 [Paenibacillus sp. JDR-2],3RDK_B Protein crystal structure of xylanase A1 of Paenibacillus sp. JDR-2 [Paenibacillus sp. JDR-2],3RO8_A Crystal structure of the catalytic domain of XynA1 from Paenibacillus sp. JDR-2 [Paenibacillus sp. JDR-2],3RO8_B Crystal structure of the catalytic domain of XynA1 from Paenibacillus sp. JDR-2 [Paenibacillus sp. JDR-2],3RO8_C Crystal structure of the catalytic domain of XynA1 from Paenibacillus sp. JDR-2 [Paenibacillus sp. JDR-2],3RO8_D Crystal structure of the catalytic domain of XynA1 from Paenibacillus sp. JDR-2 [Paenibacillus sp. JDR-2],3RO8_E Crystal structure of the catalytic domain of XynA1 from Paenibacillus sp. JDR-2 [Paenibacillus sp. JDR-2],3RO8_F Crystal structure of the catalytic domain of XynA1 from Paenibacillus sp. JDR-2 [Paenibacillus sp. JDR-2],3RO8_G Crystal structure of the catalytic domain of XynA1 from Paenibacillus sp. JDR-2 [Paenibacillus sp. JDR-2],3RO8_H Crystal structure of the catalytic domain of XynA1 from Paenibacillus sp. JDR-2 [Paenibacillus sp. JDR-2],4E4P_A Second native structure of Xylanase A1 from Paenibacillus sp. JDR-2 [Paenibacillus sp. JDR-2],4E4P_B Second native structure of Xylanase A1 from Paenibacillus sp. JDR-2 [Paenibacillus sp. JDR-2] |
| 4HU8_A | 7.33e-66 | 1311 | 1667 | 83 | 441 | CrystalStructure of a Bacterial Ig-like Domain Containing GH10 Xylanase from Termite Gut [Globitermes brachycerastes],4HU8_B Crystal Structure of a Bacterial Ig-like Domain Containing GH10 Xylanase from Termite Gut [Globitermes brachycerastes],4HU8_C Crystal Structure of a Bacterial Ig-like Domain Containing GH10 Xylanase from Termite Gut [Globitermes brachycerastes],4HU8_D Crystal Structure of a Bacterial Ig-like Domain Containing GH10 Xylanase from Termite Gut [Globitermes brachycerastes],4HU8_E Crystal Structure of a Bacterial Ig-like Domain Containing GH10 Xylanase from Termite Gut [Globitermes brachycerastes],4HU8_F Crystal Structure of a Bacterial Ig-like Domain Containing GH10 Xylanase from Termite Gut [Globitermes brachycerastes],4HU8_G Crystal Structure of a Bacterial Ig-like Domain Containing GH10 Xylanase from Termite Gut [Globitermes brachycerastes],4HU8_H Crystal Structure of a Bacterial Ig-like Domain Containing GH10 Xylanase from Termite Gut [Globitermes brachycerastes] |
| 6FHE_A | 4.30e-58 | 1328 | 1667 | 10 | 339 | Highlyactive enzymes by automated modular backbone assembly and sequence design [synthetic construct] |
| 5OFJ_A | 1.71e-56 | 1320 | 1668 | 2 | 337 | Crystalstructure of N-terminal domain of bifunctional CbXyn10C [Caldicellulosiruptor bescii DSM 6725] |
| 7NL2_A | 3.35e-56 | 1328 | 1668 | 9 | 339 | ChainA, Beta-xylanase [Pseudothermotoga thermarum DSM 5069],7NL2_B Chain B, Beta-xylanase [Pseudothermotoga thermarum DSM 5069] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| C6CRV0 | 5.33e-118 | 987 | 2363 | 189 | 1456 | Endo-1,4-beta-xylanase A OS=Paenibacillus sp. (strain JDR-2) OX=324057 GN=xynA1 PE=1 SV=1 |
| P38535 | 1.94e-72 | 1305 | 2369 | 178 | 1084 | Exoglucanase XynX OS=Acetivibrio thermocellus OX=1515 GN=xynX PE=3 SV=1 |
| Q60037 | 1.05e-57 | 1006 | 1681 | 70 | 699 | Endo-1,4-beta-xylanase A OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=xynA PE=1 SV=1 |
| Q60042 | 7.36e-57 | 1006 | 1681 | 65 | 695 | Endo-1,4-beta-xylanase A OS=Thermotoga neapolitana OX=2337 GN=xynA PE=1 SV=1 |
| P36917 | 2.75e-55 | 970 | 2292 | 21 | 1155 | Endo-1,4-beta-xylanase A OS=Thermoanaerobacterium saccharolyticum OX=28896 GN=xynA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
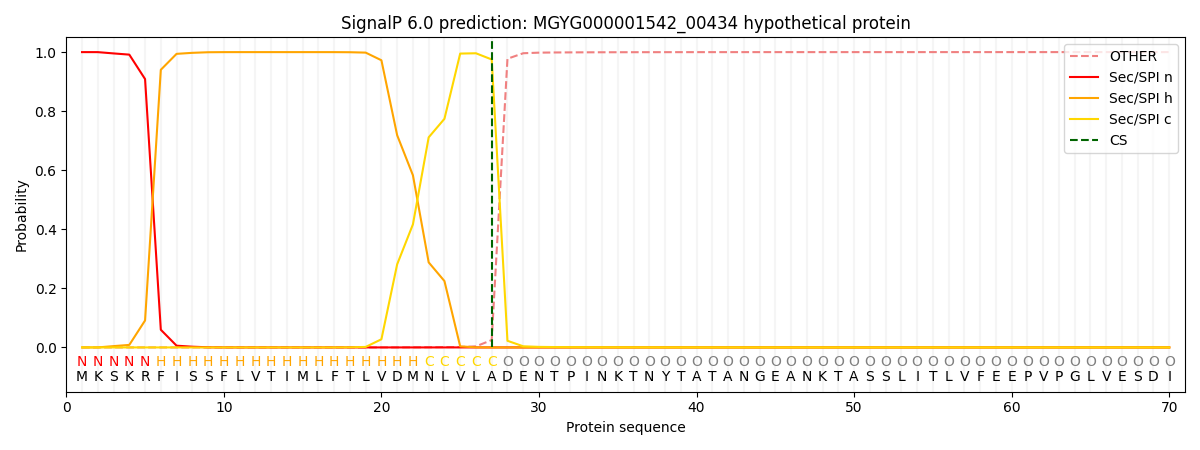
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000271 | 0.999036 | 0.000199 | 0.000171 | 0.000156 | 0.000138 |