You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001542_00914
You are here: Home > Sequence: MGYG000001542_00914
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
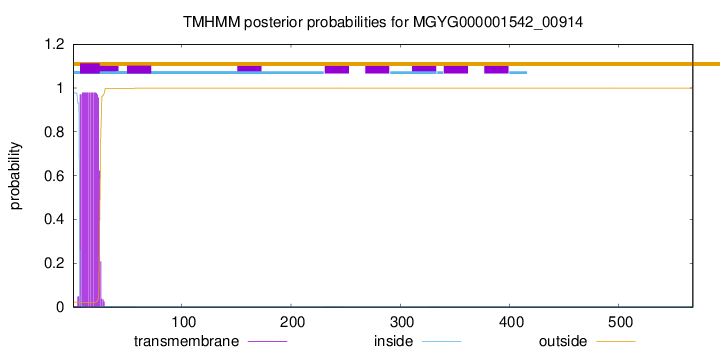
TMHMM annotations
Basic Information help
| Species | Paenibacillus_A sp900069005 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus_A; Paenibacillus_A sp900069005 | |||||||||||
| CAZyme ID | MGYG000001542_00914 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 146669; End: 148375 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 46 | 313 | 2.3e-76 | 0.9075907590759076 |
| CBM3 | 425 | 504 | 1.2e-21 | 0.9886363636363636 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3693 | XynA | 5.64e-98 | 33 | 320 | 30 | 345 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam00331 | Glyco_hydro_10 | 9.80e-87 | 33 | 313 | 16 | 309 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 2.72e-77 | 65 | 312 | 2 | 263 | Glycosyl hydrolase family 10. |
| pfam00942 | CBM_3 | 1.33e-22 | 424 | 503 | 1 | 82 | Cellulose binding domain. |
| smart01067 | CBM_3 | 1.78e-18 | 424 | 504 | 1 | 83 | Cellulose binding domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AZS17860.1 | 0.0 | 1 | 568 | 1 | 568 |
| QJD82069.1 | 3.37e-283 | 3 | 568 | 5 | 569 |
| BAK22544.1 | 6.42e-283 | 3 | 568 | 5 | 577 |
| QKS45670.1 | 1.02e-281 | 3 | 568 | 5 | 576 |
| AVI01408.1 | 1.46e-182 | 2 | 319 | 4 | 321 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4PMY_A | 4.72e-90 | 33 | 316 | 5 | 299 | Crystalstructure of GH10 endo-b-1,4-xylanase (XynB) from Xanthomonas axonopodis pv citri complexed with xylose [Xanthomonas citri pv. citri str. 306],4PMY_B Crystal structure of GH10 endo-b-1,4-xylanase (XynB) from Xanthomonas axonopodis pv citri complexed with xylose [Xanthomonas citri pv. citri str. 306],4PMZ_A Crystal structure of GH10 endo-b-1,4-xylanase (XynB) from Xanthomonas axonopodis pv citri complexed with xylobiose [Xanthomonas citri pv. citri str. 306],4PMZ_B Crystal structure of GH10 endo-b-1,4-xylanase (XynB) from Xanthomonas axonopodis pv citri complexed with xylobiose [Xanthomonas citri pv. citri str. 306] |
| 4PN2_A | 5.03e-90 | 33 | 316 | 6 | 300 | Crystalstructure of GH10 endo-b-1,4-xylanase (XynB) from Xanthomonas axonopodis pv citri complexed with xylotriose [Xanthomonas citri pv. citri str. 306],4PN2_B Crystal structure of GH10 endo-b-1,4-xylanase (XynB) from Xanthomonas axonopodis pv citri complexed with xylotriose [Xanthomonas citri pv. citri str. 306] |
| 4PMX_A | 5.20e-90 | 33 | 316 | 6 | 300 | Crystalstructure of GH10 endo-b-1,4-xylanase (XynB) from Xanthomonas axonopodis pv citri in the native form [Xanthomonas citri pv. citri str. 306] |
| 3ZQW_A | 6.64e-40 | 421 | 568 | 5 | 153 | ChainA, CELLULOSOMAL SCAFFOLDIN [Acetivibrio cellulolyticus],3ZU8_A Chain A, Cellulosomal Scaffoldin [Acetivibrio cellulolyticus],3ZUC_A Chain A, CELLULOSOMAL SCAFFOLDIN [Acetivibrio cellulolyticus] |
| 6D5B_A | 6.88e-35 | 424 | 568 | 21 | 167 | Structureof Caldicellulosiruptor danielii CBM3 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5B_B Structure of Caldicellulosiruptor danielii CBM3 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5B_C Structure of Caldicellulosiruptor danielii CBM3 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5B_D Structure of Caldicellulosiruptor danielii CBM3 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5B_E Structure of Caldicellulosiruptor danielii CBM3 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5B_F Structure of Caldicellulosiruptor danielii CBM3 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5B_G Structure of Caldicellulosiruptor danielii CBM3 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5B_H Structure of Caldicellulosiruptor danielii CBM3 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5B_I Structure of Caldicellulosiruptor danielii CBM3 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5B_J Structure of Caldicellulosiruptor danielii CBM3 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5B_K Structure of Caldicellulosiruptor danielii CBM3 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5B_L Structure of Caldicellulosiruptor danielii CBM3 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P23030 | 3.93e-64 | 37 | 312 | 323 | 592 | Endo-1,4-beta-xylanase B OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=xynB PE=1 SV=2 |
| P29718 | 2.59e-56 | 424 | 568 | 1 | 145 | Uncharacterized protein in celA 5'region (Fragment) OS=Paenibacillus lautus OX=1401 PE=4 SV=1 |
| P29719 | 9.18e-48 | 421 | 568 | 552 | 700 | Endoglucanase A OS=Paenibacillus lautus OX=1401 GN=celA PE=3 SV=1 |
| P10474 | 6.89e-47 | 54 | 568 | 76 | 570 | Endoglucanase/exoglucanase B OS=Caldicellulosiruptor saccharolyticus OX=44001 GN=celB PE=3 SV=1 |
| Q02934 | 3.00e-39 | 422 | 568 | 740 | 887 | Endoglucanase 1 OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celI PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
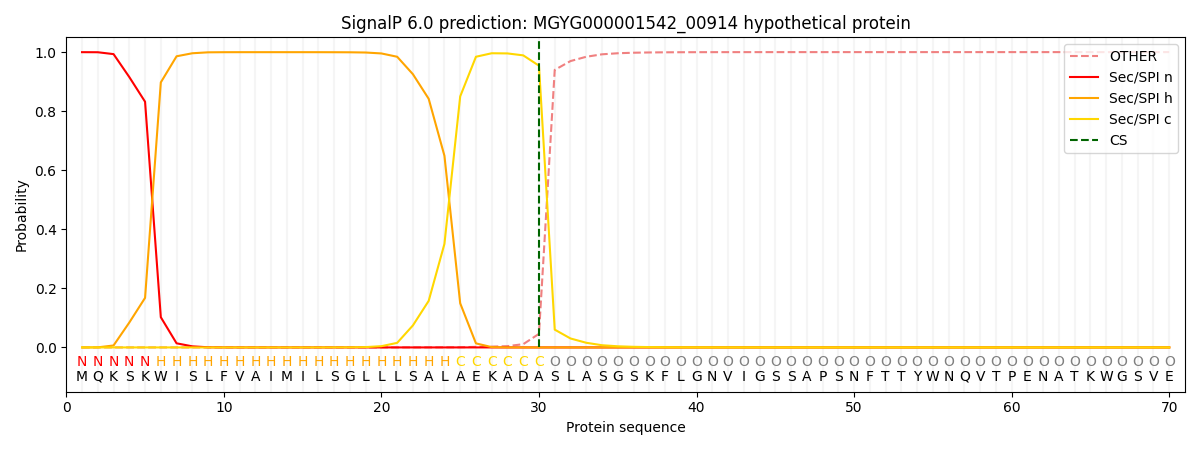
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000371 | 0.998540 | 0.000544 | 0.000198 | 0.000164 | 0.000146 |