You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001542_03476
You are here: Home > Sequence: MGYG000001542_03476
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
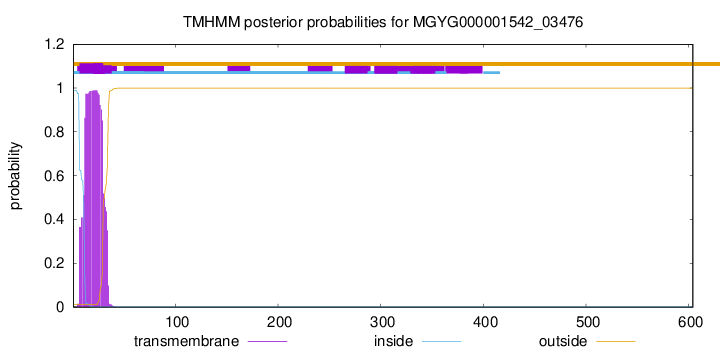
TMHMM annotations
Basic Information help
| Species | Paenibacillus_A sp900069005 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus_A; Paenibacillus_A sp900069005 | |||||||||||
| CAZyme ID | MGYG000001542_03476 | |||||||||||
| CAZy Family | PL31 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 161825; End: 163639 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL31 | 307 | 502 | 7.6e-63 | 0.9946524064171123 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| NF033679 | DNRLRE_dom | 4.85e-26 | 45 | 188 | 2 | 163 | DNRLRE domain. The DNRLRE domain, with a length of about 160 amino acids, appears typically in large, repetitive surface proteins of bacteria and archaea, sometimes repeated several times. It occurs, notably, three times in the C-terminal region of the enzyme disaggregatase from the archaeal species Methanosarcina mazei, each time with the motif DNRLRE, for which the domain is named. Archaeal proteins within this family are described particularly well by the currently more narrowly defined Pfam model, PF06848. Note that the catalytic region of disaggregatase, in the N-terminal portion of the protein, is modeled by a different HMM, PF08480. |
| cd00063 | FN3 | 1.60e-07 | 202 | 286 | 1 | 93 | Fibronectin type 3 domain; One of three types of internal repeats found in the plasma protein fibronectin. Its tenth fibronectin type III repeat contains an RGD cell recognition sequence in a flexible loop between 2 strands. Approximately 2% of all animal proteins contain the FN3 repeat; including extracellular and intracellular proteins, membrane spanning cytokine receptors, growth hormone receptors, tyrosine phosphatase receptors, and adhesion molecules. FN3-like domains are also found in bacterial glycosyl hydrolases. |
| COG3979 | COG3979 | 4.91e-05 | 198 | 377 | 1 | 181 | Chitodextrinase [Carbohydrate transport and metabolism]. |
| smart00060 | FN3 | 4.14e-04 | 202 | 273 | 1 | 81 | Fibronectin type 3 domain. One of three types of internal repeat within the plasma protein, fibronectin. The tenth fibronectin type III repeat contains a RGD cell recognition sequence in a flexible loop between 2 strands. Type III modules are present in both extracellular and intracellular proteins. |
| cd14251 | PL-6 | 4.28e-04 | 301 | 538 | 2 | 240 | Polysaccharide Lyase Family 6. Polysaccharide Lyase Family 6 is a family of beta-helical polysaccharide lyases. Members include alginate lyase (EC 4.2.2.3) and chondroitinase B (EC 4.2.2.19). Chondroitinase B is an enzyme that only cleaves the beta-(1,4)-linkage of dermatan sulfate (DS), leading to 4,5-unsaturated dermatan sulfate disaccharides as the product. DS is a highly sulfated, unbranched polysaccharide belonging to a family of glycosaminoglycans (GAGs) composed of alternating hexosamine (gluco- or galactosamine) and uronic acid (D-glucuronic or L-iduronic acid) moieties. DS contains alternating 1,4-beta-D-galactosamine (GalNac) and 1,3-alpha-L-iduronic acid units. The related chondroitin sulfate (CS) contains alternating GalNac and 1,3-beta-D-glucuronic acid units. Alginate lyases (known as either mannuronate (EC 4.2.2.3) or guluronate lyases (EC 4.2.2.11) catalyze the degradation of alginate, a copolymer of alpha-L-guluronate and its C5 epimer beta-D-mannuronate. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AZS15743.1 | 0.0 | 1 | 604 | 1 | 604 |
| QRN99389.1 | 1.02e-259 | 39 | 604 | 41 | 604 |
| ADO69469.1 | 1.39e-259 | 40 | 603 | 42 | 602 |
| QRK13860.1 | 1.52e-255 | 40 | 604 | 39 | 599 |
| ATB42714.1 | 4.21e-246 | 39 | 604 | 49 | 611 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6KFN_A | 2.23e-80 | 302 | 603 | 10 | 298 | Crystalstructure of alginate lyase from Paenibacillus sp. str. FPU-7 [Paenibacillus sp. FPU-7] |
| 1K85_A | 2.76e-11 | 202 | 286 | 4 | 88 | ChainA, CHITINASE A1 [Niallia circulans] |
| 1RU4_A | 1.55e-10 | 270 | 440 | 2 | 171 | ChainA, Pectate lyase [Dickeya chrysanthemi] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P20533 | 2.72e-13 | 127 | 303 | 475 | 661 | Chitinase A1 OS=Niallia circulans OX=1397 GN=chiA1 PE=1 SV=1 |
| P0C1A6 | 1.77e-11 | 266 | 440 | 23 | 196 | Pectate lyase L OS=Dickeya chrysanthemi OX=556 GN=pelL PE=3 SV=1 |
| P0C1A7 | 7.31e-11 | 266 | 440 | 23 | 196 | Pectate lyase L OS=Dickeya dadantii (strain 3937) OX=198628 GN=pelL PE=1 SV=1 |
| A0P8X0 | 1.65e-08 | 83 | 286 | 690 | 899 | Alpha-amylase OS=Niallia circulans OX=1397 GN=igtZ PE=1 SV=1 |
| P50899 | 2.74e-08 | 181 | 289 | 777 | 887 | Exoglucanase B OS=Cellulomonas fimi (strain ATCC 484 / DSM 20113 / JCM 1341 / NBRC 15513 / NCIMB 8980 / NCTC 7547) OX=590998 GN=cbhB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
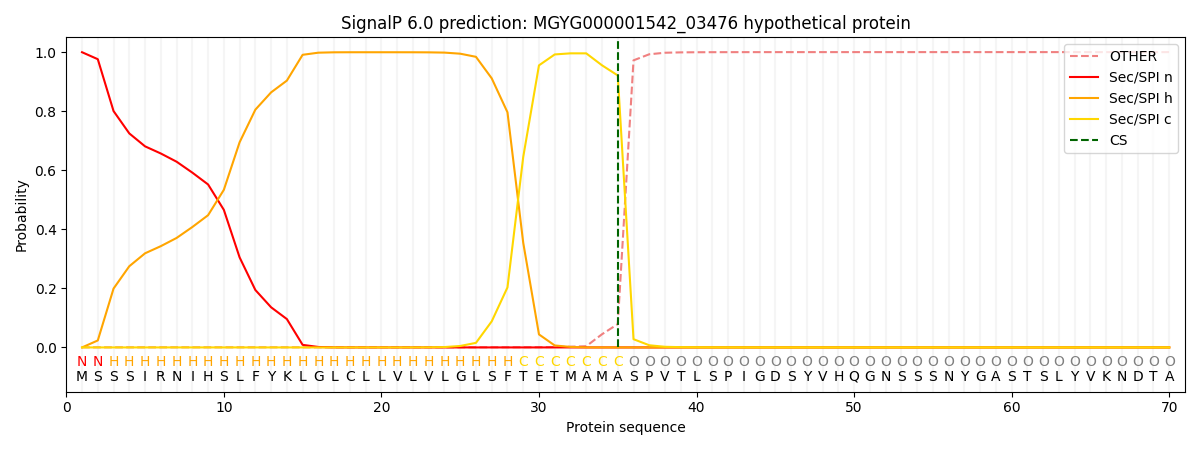
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001155 | 0.998008 | 0.000259 | 0.000191 | 0.000175 | 0.000184 |