You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001542_04878
You are here: Home > Sequence: MGYG000001542_04878
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paenibacillus_A sp900069005 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus_A; Paenibacillus_A sp900069005 | |||||||||||
| CAZyme ID | MGYG000001542_04878 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 660468; End: 664223 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 71 | 331 | 5.9e-88 | 0.9915611814345991 |
| CBM17 | 364 | 564 | 8e-82 | 0.9950738916256158 |
| CBM28 | 581 | 753 | 2.9e-64 | 0.8461538461538461 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam03424 | CBM_17_28 | 1.94e-92 | 364 | 565 | 1 | 204 | Carbohydrate binding domain (family 17/28). |
| pfam00150 | Cellulase | 2.40e-78 | 69 | 338 | 2 | 272 | Cellulase (glycosyl hydrolase family 5). |
| NF033190 | inl_like_NEAT_1 | 2.07e-16 | 1071 | 1243 | 580 | 746 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
| pfam00395 | SLH | 1.56e-10 | 1134 | 1175 | 1 | 42 | S-layer homology domain. |
| COG2730 | BglC | 8.42e-09 | 102 | 344 | 83 | 372 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AAT97264.1 | 0.0 | 1 | 753 | 1 | 758 |
| BAA12744.1 | 0.0 | 3 | 753 | 4 | 760 |
| ACW22975.1 | 0.0 | 3 | 765 | 4 | 771 |
| CAT16607.1 | 0.0 | 3 | 765 | 4 | 771 |
| ABH71811.1 | 0.0 | 3 | 765 | 4 | 771 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5ECU_A | 5.69e-196 | 49 | 754 | 20 | 543 | Theunliganded structure of Caldicellulosiruptor saccharolyticus GH5 [Caldicellulosiruptor saccharolyticus] |
| 1G01_A | 3.00e-166 | 50 | 397 | 12 | 364 | AlkalineCellulase K Catalytic Domain [Bacillus sp. KSM-635],1G0C_A ALKALINE CELLULASE K CATALYTIC DOMAIN-CELLOBIOSE COMPLEX [Bacillus sp. KSM-635] |
| 5FIP_A | 1.06e-147 | 49 | 391 | 4 | 331 | Discoveryand characterization of a novel thermostable and highly halotolerant GH5 cellulase from an Icelandic hot spring isolate [unidentified],5FIP_B Discovery and characterization of a novel thermostable and highly halotolerant GH5 cellulase from an Icelandic hot spring isolate [unidentified],5FIP_C Discovery and characterization of a novel thermostable and highly halotolerant GH5 cellulase from an Icelandic hot spring isolate [unidentified],5FIP_D Discovery and characterization of a novel thermostable and highly halotolerant GH5 cellulase from an Icelandic hot spring isolate [unidentified] |
| 6GJF_A | 4.95e-80 | 67 | 382 | 20 | 301 | Ancestralendocellulase Cel5A [synthetic construct],6GJF_B Ancestral endocellulase Cel5A [synthetic construct],6GJF_C Ancestral endocellulase Cel5A [synthetic construct],6GJF_D Ancestral endocellulase Cel5A [synthetic construct],6GJF_E Ancestral endocellulase Cel5A [synthetic construct],6GJF_F Ancestral endocellulase Cel5A [synthetic construct] |
| 1UWW_A | 1.03e-79 | 566 | 753 | 1 | 188 | X-raycrystal structure of a non-crystalline cellulose specific carbohydrate-binding module: CBM28. [Alkalihalobacillus akibai],1UWW_B X-ray crystal structure of a non-crystalline cellulose specific carbohydrate-binding module: CBM28. [Alkalihalobacillus akibai] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P19424 | 0.0 | 50 | 753 | 232 | 940 | Endoglucanase OS=Bacillus sp. (strain KSM-635) OX=1415 PE=1 SV=1 |
| P06564 | 0.0 | 3 | 765 | 4 | 769 | Endoglucanase OS=Alkalihalobacillus akibai (strain ATCC 43226 / DSM 21942 / CIP 109018 / JCM 9157 / 1139) OX=1236973 PE=1 SV=1 |
| P19570 | 4.59e-313 | 1 | 752 | 1 | 794 | Endoglucanase C OS=Evansella cellulosilytica (strain ATCC 21833 / DSM 2522 / FERM P-1141 / JCM 9156 / N-4) OX=649639 GN=celC PE=3 SV=1 |
| A0A0U4EBH5 | 3.48e-146 | 49 | 391 | 53 | 380 | Cellulase CelDZ1 OS=Thermoanaerobacterium sp. OX=40549 GN=celDZ1a PE=1 SV=1 |
| O85465 | 2.93e-75 | 67 | 388 | 45 | 333 | Endoglucanase 5A OS=Salipaludibacillus agaradhaerens OX=76935 GN=cel5A PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
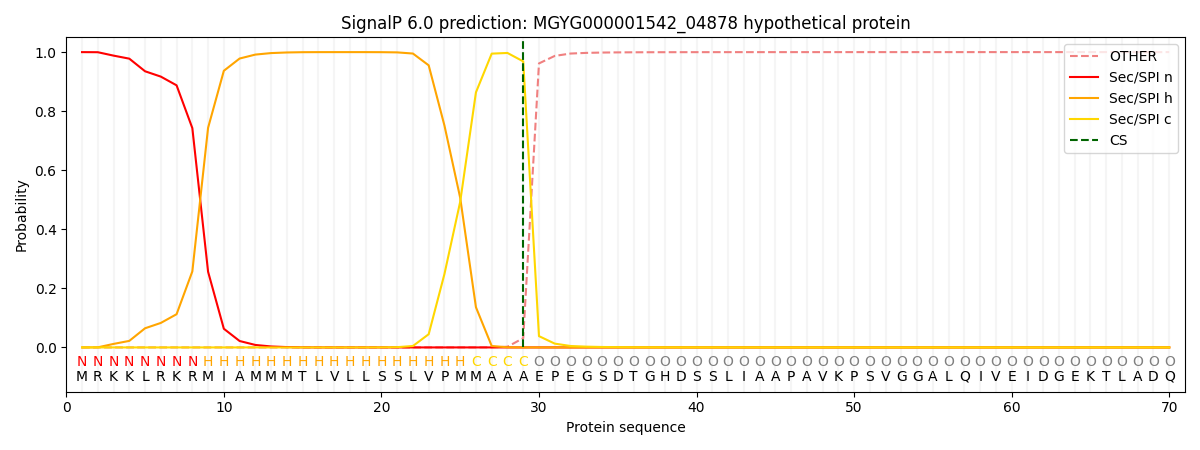
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000313 | 0.998904 | 0.000223 | 0.000211 | 0.000164 | 0.000142 |
