You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001543_02419
You are here: Home > Sequence: MGYG000001543_02419
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
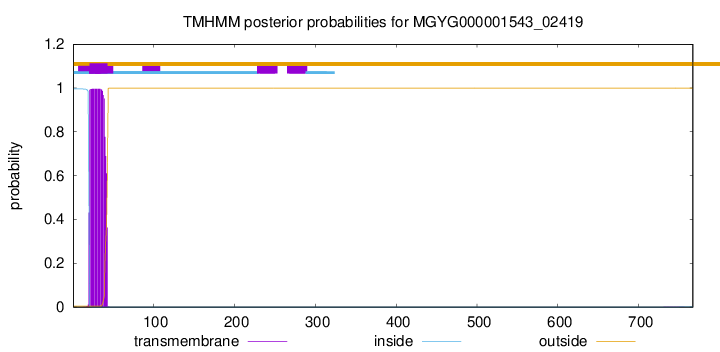
TMHMM annotations
Basic Information help
| Species | Lachnoclostridium sp900078195 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Lachnoclostridium; Lachnoclostridium sp900078195 | |||||||||||
| CAZyme ID | MGYG000001543_02419 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 863481; End: 865784 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH18 | 100 | 427 | 9.5e-27 | 0.8682432432432432 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| sd00033 | LRR_RI | 5.25e-43 | 515 | 747 | 1 | 229 | leucine-rich repeats, ribonuclease inhibitor (RI)-like subfamily. A leucine-rich repeat (LRR) is a structural protein motif of 20-30 amino acids that is unusually rich in the hydrophobic amino acid leucine. The conserved eleven-residue sequence motif (LxxLxLxxN/CxL) within the LRRs corresponds to the beta-strand and adjacent loop regions, whereas the remaining parts of the repeats are variable. LRRs fold together to form a solenoid protein domain, termed leucine-rich repeat domain. Leucine-rich repeats are usually involved in protein-protein interactions. |
| NF033189 | internalin_A | 3.02e-39 | 466 | 754 | 47 | 315 | class 1 internalin InlA. Internalins, as found in the intracellular human pathogen Listeria monocytogenes, are paralogous surface or secreted proteins with an N-terminal signal peptide, leucine-rich repeats, and usually a C-terminal LPXTG processing and cell surface anchoring site. See PMID:17764999 for a general discussion of internalins. Members of this family are internalin A (InlA), a class 1 (LPXTG-type) internalin. |
| sd00033 | LRR_RI | 5.82e-38 | 509 | 732 | 19 | 238 | leucine-rich repeats, ribonuclease inhibitor (RI)-like subfamily. A leucine-rich repeat (LRR) is a structural protein motif of 20-30 amino acids that is unusually rich in the hydrophobic amino acid leucine. The conserved eleven-residue sequence motif (LxxLxLxxN/CxL) within the LRRs corresponds to the beta-strand and adjacent loop regions, whereas the remaining parts of the repeats are variable. LRRs fold together to form a solenoid protein domain, termed leucine-rich repeat domain. Leucine-rich repeats are usually involved in protein-protein interactions. |
| NF033189 | internalin_A | 6.88e-38 | 491 | 720 | 184 | 413 | class 1 internalin InlA. Internalins, as found in the intracellular human pathogen Listeria monocytogenes, are paralogous surface or secreted proteins with an N-terminal signal peptide, leucine-rich repeats, and usually a C-terminal LPXTG processing and cell surface anchoring site. See PMID:17764999 for a general discussion of internalins. Members of this family are internalin A (InlA), a class 1 (LPXTG-type) internalin. |
| NF033189 | internalin_A | 1.08e-36 | 491 | 749 | 140 | 398 | class 1 internalin InlA. Internalins, as found in the intracellular human pathogen Listeria monocytogenes, are paralogous surface or secreted proteins with an N-terminal signal peptide, leucine-rich repeats, and usually a C-terminal LPXTG processing and cell surface anchoring site. See PMID:17764999 for a general discussion of internalins. Members of this family are internalin A (InlA), a class 1 (LPXTG-type) internalin. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ABX42184.1 | 4.27e-256 | 2 | 527 | 5 | 532 |
| AEY67763.1 | 7.86e-149 | 66 | 525 | 38 | 481 |
| ACL77187.1 | 1.53e-145 | 60 | 525 | 32 | 481 |
| AOT70741.1 | 6.03e-64 | 66 | 447 | 88 | 469 |
| AUS96550.1 | 3.18e-52 | 62 | 449 | 216 | 587 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
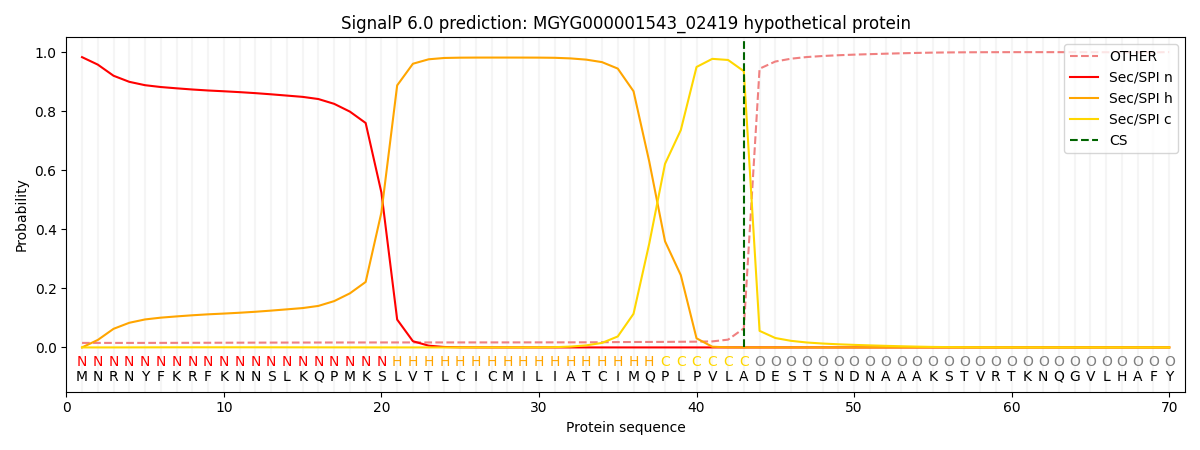
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.017819 | 0.978748 | 0.002526 | 0.000335 | 0.000270 | 0.000267 |