You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001543_02878
You are here: Home > Sequence: MGYG000001543_02878
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Lachnoclostridium sp900078195 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Lachnoclostridium; Lachnoclostridium sp900078195 | |||||||||||
| CAZyme ID | MGYG000001543_02878 | |||||||||||
| CAZy Family | CBM22 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1444367; End: 1446670 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 206 | 551 | 3.3e-93 | 0.9933993399339934 |
| CBM22 | 43 | 171 | 6.4e-34 | 0.9541984732824428 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00331 | Glyco_hydro_10 | 2.38e-114 | 207 | 551 | 1 | 310 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 1.12e-99 | 274 | 549 | 12 | 263 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 5.59e-72 | 210 | 551 | 34 | 339 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam02018 | CBM_4_9 | 3.08e-24 | 39 | 171 | 1 | 130 | Carbohydrate binding domain. This family includes diverse carbohydrate binding domains. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ABX41011.1 | 0.0 | 1 | 767 | 1 | 756 |
| BCJ96140.1 | 0.0 | 1 | 766 | 1 | 730 |
| CUH92215.1 | 0.0 | 3 | 767 | 2 | 707 |
| ACZ98623.1 | 1.54e-305 | 2 | 767 | 6 | 699 |
| BAA21516.2 | 1.62e-277 | 1 | 558 | 1 | 550 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2W5F_A | 4.57e-103 | 41 | 549 | 25 | 524 | ChainA, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],2W5F_B Chain B, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus] |
| 2WYS_A | 2.15e-99 | 41 | 549 | 25 | 524 | ChainA, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],2WYS_B Chain B, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],2WZE_A Chain A, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],2WZE_B Chain B, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus] |
| 5OFJ_A | 4.90e-70 | 203 | 551 | 7 | 337 | Crystalstructure of N-terminal domain of bifunctional CbXyn10C [Caldicellulosiruptor bescii DSM 6725] |
| 5OFK_A | 3.58e-69 | 203 | 551 | 7 | 337 | Crystalstructure of CbXyn10C variant E140Q/E248Q complexed with xyloheptaose [Caldicellulosiruptor bescii DSM 6725],5OFL_A Crystal structure of CbXyn10C variant E140Q/E248Q complexed with cellohexaose [Caldicellulosiruptor bescii DSM 6725] |
| 6D5C_A | 2.61e-67 | 203 | 551 | 19 | 349 | Structureof Caldicellulosiruptor danielii GH10 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5C_B Structure of Caldicellulosiruptor danielii GH10 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5C_C Structure of Caldicellulosiruptor danielii GH10 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P51584 | 4.37e-98 | 41 | 549 | 36 | 535 | Endo-1,4-beta-xylanase Y OS=Acetivibrio thermocellus OX=1515 GN=xynY PE=1 SV=1 |
| Q60042 | 2.05e-94 | 41 | 557 | 200 | 695 | Endo-1,4-beta-xylanase A OS=Thermotoga neapolitana OX=2337 GN=xynA PE=1 SV=1 |
| Q60037 | 5.75e-94 | 41 | 557 | 205 | 699 | Endo-1,4-beta-xylanase A OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=xynA PE=1 SV=1 |
| P29126 | 1.23e-74 | 206 | 550 | 629 | 950 | Bifunctional endo-1,4-beta-xylanase XylA OS=Ruminococcus flavefaciens OX=1265 GN=xynA PE=3 SV=1 |
| P36917 | 1.30e-74 | 39 | 559 | 195 | 682 | Endo-1,4-beta-xylanase A OS=Thermoanaerobacterium saccharolyticum OX=28896 GN=xynA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
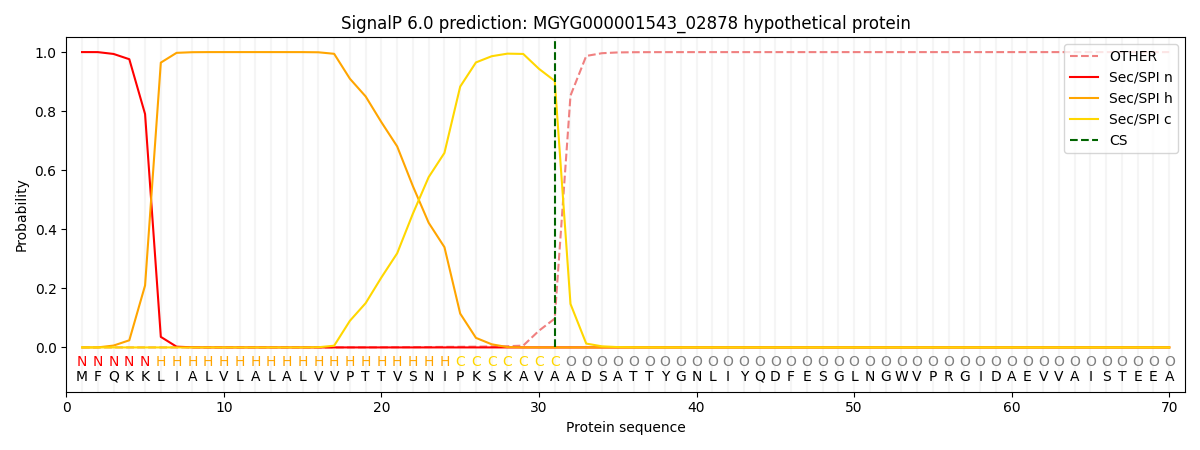
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000292 | 0.998994 | 0.000203 | 0.000165 | 0.000162 | 0.000148 |
