You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001545_00393
You are here: Home > Sequence: MGYG000001545_00393
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
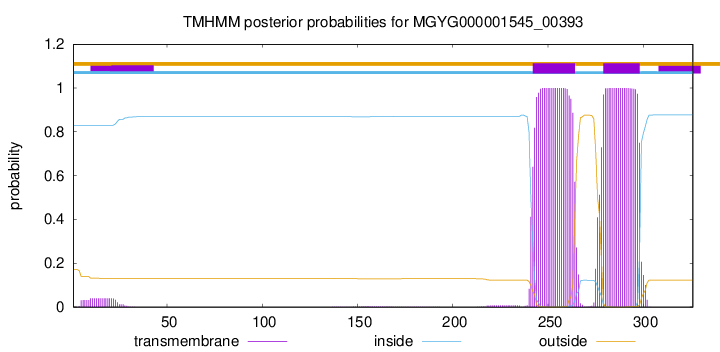
TMHMM annotations
Basic Information help
| Species | Olegusella massiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Coriobacteriia; Coriobacteriales; Atopobiaceae; Olegusella; Olegusella massiliensis | |||||||||||
| CAZyme ID | MGYG000001545_00393 | |||||||||||
| CAZy Family | GT81 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 369725; End: 370705 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT81 | 16 | 210 | 1.2e-20 | 0.7133105802047781 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| TIGR04182 | glyco_TIGR04182 | 1.70e-82 | 19 | 306 | 5 | 287 | glycosyltransferase, TIGR04182 family. Members of this family are glycosyltransferases restricted to the archaea. All but two members are from species with the PGF-CTERM/archaeosortase A system, a proposed maturation system for exported, glycosylated proteins as are found often in S-layers. |
| cd04179 | DPM_DPG-synthase_like | 2.20e-42 | 19 | 193 | 2 | 185 | DPM_DPG-synthase_like is a member of the Glycosyltransferase 2 superfamily. DPM1 is the catalytic subunit of eukaryotic dolichol-phosphate mannose (DPM) synthase. DPM synthase is required for synthesis of the glycosylphosphatidylinositol (GPI) anchor, N-glycan precursor, protein O-mannose, and C-mannose. In higher eukaryotes,the enzyme has three subunits, DPM1, DPM2 and DPM3. DPM is synthesized from dolichol phosphate and GDP-Man on the cytosolic surface of the ER membrane by DPM synthase and then is flipped onto the luminal side and used as a donor substrate. In lower eukaryotes, such as Saccharomyces cerevisiae and Trypanosoma brucei, DPM synthase consists of a single component (Dpm1p and TbDpm1, respectively) that possesses one predicted transmembrane region near the C terminus for anchoring to the ER membrane. In contrast, the Dpm1 homologues of higher eukaryotes, namely fission yeast, fungi, and animals, have no transmembrane region, suggesting the existence of adapter molecules for membrane anchoring. This family also includes bacteria and archaea DPM1_like enzymes. However, the enzyme structure and mechanism of function are not well understood. The UDP-glucose:dolichyl-phosphate glucosyltransferase (DPG_synthase) is a transmembrane-bound enzyme of the endoplasmic reticulum involved in protein N-linked glycosylation. This enzyme catalyzes the transfer of glucose from UDP-glucose to dolichyl phosphate. This protein family belongs to Glycosyltransferase 2 superfamily. |
| 133062 | cd06442 | 6.87e-25 | 19 | 219 | 2 | 210 | DPM1_like DPM1_like represents putative enzymes similar to eukaryotic DPM1. Proteins similar to eukaryotic DPM1, including enzymes from bacteria and archaea; DPM1 is the catalytic subunit of eukaryotic dolichol-phosphate mannose (DPM) synthase. DPM synthase is required for synthesis of the glycosylphosphatidylinositol (GPI) anchor, N-glycan precursor, protein O-mannose, and C-mannose. In higher eukaryotes,the enzyme has three subunits, DPM1, DPM2 and DPM3. DPM is synthesized from dolichol phosphate and GDP-Man on the cytosolic surface of the ER membrane by DPM synthase and then is flipped onto the luminal side and used as a donor substrate. In lower eukaryotes, such as Saccharomyces cerevisiae and Trypanosoma brucei, DPM synthase consists of a single component (Dpm1p and TbDpm1, respectively) that possesses one predicted transmembrane region near the C terminus for anchoring to the ER membrane. In contrast, the Dpm1 homologues of higher eukaryotes, namely fission yeast, fungi, and animals, have no transmembrane region, suggesting the existence of adapter molecules for membrane anchoring. This family also includes bacteria and archaea DPM1_like enzymes. However, the enzyme structure and mechanism of function are not well understood. This protein family belongs to Glycosyltransferase 2 superfamily. |
| cd04188 | DPG_synthase | 1.10e-20 | 20 | 214 | 3 | 210 | DPG_synthase is involved in protein N-linked glycosylation. UDP-glucose:dolichyl-phosphate glucosyltransferase (DPG_synthase) is a transmembrane-bound enzyme of the endoplasmic reticulum involved in protein N-linked glycosylation. This enzyme catalyzes the transfer of glucose from UDP-glucose to dolichyl phosphate. |
| pfam00535 | Glycos_transf_2 | 1.03e-15 | 19 | 170 | 3 | 161 | Glycosyl transferase family 2. Diverse family, transferring sugar from UDP-glucose, UDP-N-acetyl- galactosamine, GDP-mannose or CDP-abequose, to a range of substrates including cellulose, dolichol phosphate and teichoic acids. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AKT49514.1 | 1.03e-153 | 16 | 326 | 11 | 321 |
| ACV55215.1 | 2.64e-148 | 16 | 319 | 15 | 318 |
| BAK44347.1 | 2.86e-147 | 14 | 321 | 10 | 317 |
| QOS69497.1 | 3.41e-147 | 16 | 319 | 17 | 320 |
| ANE22624.1 | 5.89e-144 | 12 | 319 | 16 | 323 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P37786 | 1.30e-35 | 16 | 135 | 7 | 126 | Protein RfbJ OS=Shigella flexneri OX=623 GN=rfbJ PE=3 SV=2 |
| D4GYG3 | 6.36e-35 | 10 | 306 | 1 | 293 | Glycosyltransferase AglJ OS=Haloferax volcanii (strain ATCC 29605 / DSM 3757 / JCM 8879 / NBRC 14742 / NCIMB 2012 / VKM B-1768 / DS2) OX=309800 GN=aglJ PE=1 SV=1 |
| P37787 | 2.40e-31 | 161 | 325 | 1 | 161 | Uncharacterized protein SF2094/S2216 OS=Shigella flexneri OX=623 GN=SF2094 PE=4 SV=1 |
| D4GU74 | 7.32e-20 | 12 | 224 | 42 | 255 | Low-salt glycan biosynthesis hexosyltransferase Agl6 OS=Haloferax volcanii (strain ATCC 29605 / DSM 3757 / JCM 8879 / NBRC 14742 / NCIMB 2012 / VKM B-1768 / DS2) OX=309800 GN=agl6 PE=3 SV=1 |
| B3VA58 | 7.44e-10 | 13 | 205 | 3 | 216 | UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminyltransferase OS=Methanococcus voltae OX=2188 GN=aglK PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
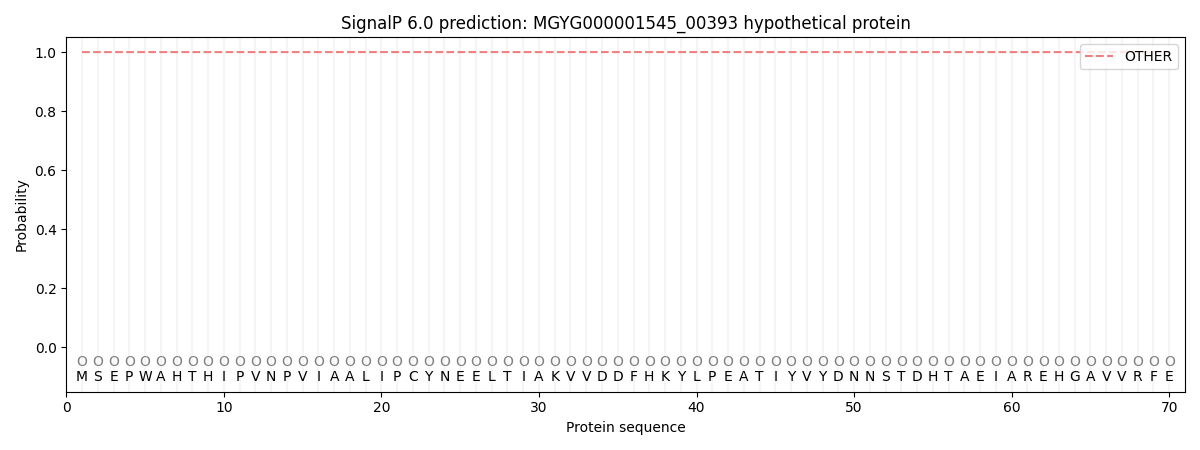
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000047 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |