You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001546_00034
You are here: Home > Sequence: MGYG000001546_00034
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes provencensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; Alistipes provencensis | |||||||||||
| CAZyme ID | MGYG000001546_00034 | |||||||||||
| CAZy Family | CE1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 20417; End: 21262 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE1 | 31 | 273 | 2e-60 | 0.9691629955947136 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0627 | FrmB | 7.55e-34 | 41 | 281 | 38 | 314 | S-formylglutathione hydrolase FrmB [Defense mechanisms]. |
| COG2382 | Fes | 4.70e-29 | 30 | 274 | 74 | 294 | Enterochelin esterase or related enzyme [Inorganic ion transport and metabolism]. |
| pfam00756 | Esterase | 1.80e-28 | 31 | 274 | 1 | 245 | Putative esterase. This family contains Esterase D. However it is not clear if all members of the family have the same function. This family is related to the pfam00135 family. |
| PRK10566 | PRK10566 | 3.71e-08 | 95 | 208 | 57 | 174 | esterase; Provisional |
| COG4099 | COG4099 | 3.41e-07 | 35 | 178 | 171 | 302 | Predicted peptidase [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QJW91289.1 | 3.22e-40 | 30 | 271 | 145 | 384 |
| VTQ01161.1 | 9.62e-38 | 30 | 270 | 41 | 275 |
| AHF26056.1 | 1.23e-37 | 14 | 270 | 130 | 396 |
| AMQ56447.1 | 6.53e-37 | 31 | 268 | 150 | 377 |
| VEH16110.1 | 2.26e-36 | 31 | 272 | 157 | 393 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5VOL_A | 2.09e-76 | 21 | 281 | 9 | 274 | Bacint_04212ferulic acid esterase [Bacteroides intestinalis DSM 17393],5VOL_B Bacint_04212 ferulic acid esterase [Bacteroides intestinalis DSM 17393],5VOL_C Bacint_04212 ferulic acid esterase [Bacteroides intestinalis DSM 17393],5VOL_D Bacint_04212 ferulic acid esterase [Bacteroides intestinalis DSM 17393],5VOL_E Bacint_04212 ferulic acid esterase [Bacteroides intestinalis DSM 17393],5VOL_F Bacint_04212 ferulic acid esterase [Bacteroides intestinalis DSM 17393],5VOL_G Bacint_04212 ferulic acid esterase [Bacteroides intestinalis DSM 17393],5VOL_H Bacint_04212 ferulic acid esterase [Bacteroides intestinalis DSM 17393] |
| 6RZN_A | 2.76e-33 | 31 | 270 | 139 | 379 | Crystalstructure of the N-terminal carbohydrate binding module family 48 and ferulic acid esterase from the multi-enzyme CE1-GH62-GH10 [uncultured bacterium],6RZN_B Crystal structure of the N-terminal carbohydrate binding module family 48 and ferulic acid esterase from the multi-enzyme CE1-GH62-GH10 [uncultured bacterium] |
| 4RGY_A | 2.93e-22 | 31 | 275 | 11 | 261 | Structuraland functional analysis of a low-temperature-active alkaline esterase from South China Sea marine sediment microbial metagenomic library [uncultured bacterium FLS12],4RGY_B Structural and functional analysis of a low-temperature-active alkaline esterase from South China Sea marine sediment microbial metagenomic library [uncultured bacterium FLS12] |
| 1JJF_A | 1.94e-20 | 42 | 272 | 50 | 254 | ChainA, Endo-1,4-beta-xylanase Z [Acetivibrio thermocellus] |
| 1JT2_A | 5.14e-20 | 42 | 272 | 50 | 254 | ChainA, PROTEIN (ENDO-1,4-BETA-XYLANASE Z) [Acetivibrio thermocellus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| D5EXZ4 | 2.83e-19 | 42 | 272 | 449 | 665 | Carbohydrate acetyl esterase/feruloyl esterase OS=Prevotella ruminicola (strain ATCC 19189 / JCM 8958 / 23) OX=264731 GN=axe1-6A PE=1 SV=1 |
| P10478 | 1.02e-18 | 42 | 272 | 69 | 273 | Endo-1,4-beta-xylanase Z OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=xynZ PE=1 SV=3 |
| D5EY13 | 1.32e-18 | 31 | 178 | 507 | 654 | Endo-1,4-beta-xylanase/feruloyl esterase OS=Prevotella ruminicola (strain ATCC 19189 / JCM 8958 / 23) OX=264731 GN=xyn10D-fae1A PE=1 SV=1 |
| P9WM38 | 2.69e-14 | 27 | 178 | 177 | 319 | Esterase MT1326 OS=Mycobacterium tuberculosis (strain CDC 1551 / Oshkosh) OX=83331 GN=MT1326 PE=3 SV=1 |
| P9WM39 | 2.69e-14 | 27 | 178 | 177 | 319 | Esterase Rv1288 OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=Rv1288 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
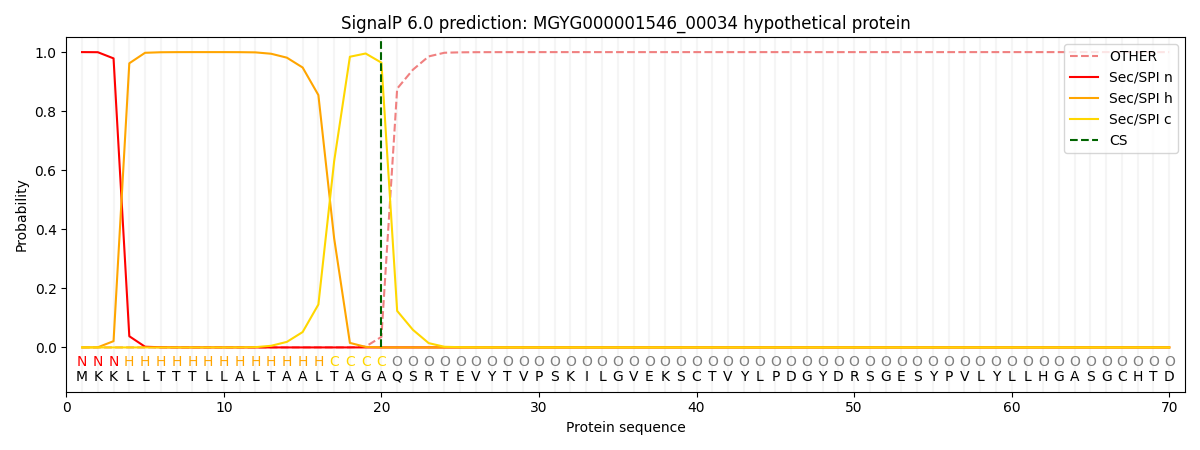
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000289 | 0.999039 | 0.000183 | 0.000174 | 0.000151 | 0.000132 |
