You are browsing environment: HUMAN GUT
MGYG000001546_00394
Basic Information
help
Species
Alistipes provencensis
Lineage
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; Alistipes provencensis
CAZyme ID
MGYG000001546_00394
CAZy Family
GT22
CAZyme Description
hypothetical protein
CAZyme Property
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000001546
3805103
Isolate
not provided
not provided
Gene Location
Start: 454592;
End: 456139
Strand: -
No EC number prediction in MGYG000001546_00394.
Family
Start
End
Evalue
family coverage
GT22
20
370
5.2e-49
0.8637532133676092
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam03901
Glyco_transf_22
5.89e-11
24
319
11
312
Alg9-like mannosyltransferase family. Members of this family are mannosyltransferase enzymes. At least some members are localized in endoplasmic reticulum and involved in GPI anchor biosynthesis.
more
PLN02816
PLN02816
1.28e-08
33
319
60
339
mannosyltransferase
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
Q94A15
1.30e-10
27
327
53
347
Mannosyltransferase APTG1 OS=Arabidopsis thaliana OX=3702 GN=APTG1 PE=2 SV=1
more
Q9JJQ0
6.90e-06
35
327
73
356
GPI mannosyltransferase 3 OS=Mus musculus OX=10090 GN=Pigb PE=2 SV=2
more
This protein is predicted as OTHER
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
1.000047
0.000001
0.000000
0.000000
0.000000
0.000000
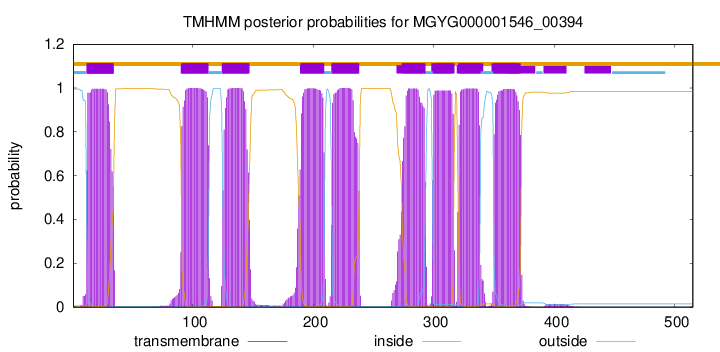
start
end
12
34
91
113
125
147
190
209
216
238
274
293
300
317
321
338
350
372