You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001546_00594
You are here: Home > Sequence: MGYG000001546_00594
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes provencensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; Alistipes provencensis | |||||||||||
| CAZyme ID | MGYG000001546_00594 | |||||||||||
| CAZy Family | GH39 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 702128; End: 703600 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH39 | 95 | 411 | 1.1e-33 | 0.7587006960556845 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01229 | Glyco_hydro_39 | 4.24e-12 | 101 | 356 | 74 | 347 | Glycosyl hydrolases family 39. |
| COG2723 | BglB | 7.71e-10 | 79 | 176 | 70 | 170 | Beta-glucosidase/6-phospho-beta-glucosidase/beta-galactosidase [Carbohydrate transport and metabolism]. |
| COG3693 | XynA | 3.32e-07 | 69 | 175 | 47 | 151 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam02449 | Glyco_hydro_42 | 4.24e-07 | 79 | 175 | 21 | 138 | Beta-galactosidase. This group of beta-galactosidase enzymes belong to the glycosyl hydrolase 42 family. The enzyme catalyzes the hydrolysis of terminal, non-reducing terminal beta-D-galactosidase residues. |
| smart00633 | Glyco_10 | 1.74e-04 | 95 | 201 | 7 | 120 | Glycosyl hydrolase family 10. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBL15167.1 | 2.67e-225 | 22 | 490 | 26 | 496 |
| BBL03131.1 | 1.08e-224 | 22 | 490 | 26 | 496 |
| QNN22472.1 | 1.07e-158 | 7 | 478 | 12 | 477 |
| QNN22723.1 | 5.60e-109 | 36 | 473 | 34 | 475 |
| ANB10603.1 | 6.12e-94 | 31 | 472 | 218 | 662 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4EKJ_A | 1.18e-07 | 74 | 243 | 48 | 231 | ChainA, Beta-xylosidase [Caulobacter vibrioides] |
| 4M29_A | 1.18e-07 | 74 | 243 | 48 | 231 | Structureof a GH39 Beta-xylosidase from Caulobacter crescentus [Caulobacter vibrioides CB15] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
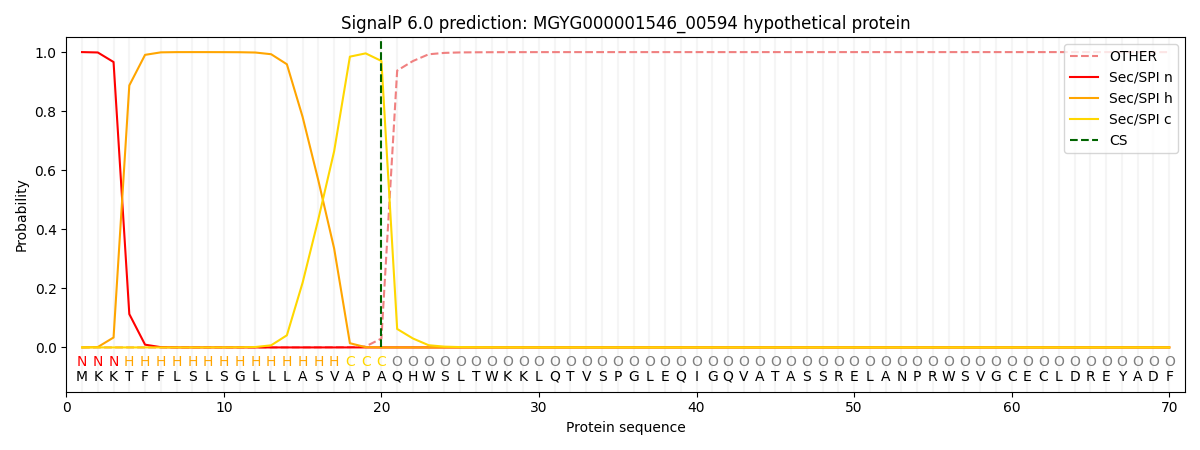
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000605 | 0.998642 | 0.000213 | 0.000192 | 0.000177 | 0.000173 |
