You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001548_02650
You are here: Home > Sequence: MGYG000001548_02650
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paenibacillus_A tuaregi | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus_A; Paenibacillus_A tuaregi | |||||||||||
| CAZyme ID | MGYG000001548_02650 | |||||||||||
| CAZy Family | CBM56 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2814351; End: 2815694 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH64 | 139 | 357 | 2.3e-57 | 0.5831062670299727 |
| CBM56 | 22 | 118 | 1.1e-30 | 0.5974842767295597 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd09214 | GH64-like | 7.61e-139 | 141 | 446 | 1 | 319 | glycosyl hydrolase 64 family. This family is represented by the laminaripentaose-producing, beta-1,3-glucanase (LPHase) of Streptomyces matensis and related bacterial and ascomycete proteins. LPHase is a member of glycoside hydrolase family 64 (GH64), it is an inverting enzyme involved in the cleavage of long-chain polysaccharide beta-1,3-glucans, into specific pentasaccharide oligomers. LPHase is a two-domain crescent fold structure: one domain is composed of 10 beta-strands, eight coming from the N-terminus of the protein and two from the C-terminal region, and the protein has a second inserted domain; this cd includes both domains. This protein has an electronegative, substrate-binding cleft, and conserved Glu and Asp residues involved in the cleavage of the beta-1,3-glucan, laminarin, a plant and fungal cell wall component. Among bacteria, many beta-1,3-glucanases are implicated in fungal cell wall degradation. Also included in this family is GluB , the beta-1,3-glucanase B from Lysobacter enzymogenes Strain N4-7. Recombinant GluB demonstrated higher relative activity toward the branched-chain beta-1,3 glucan substrate zymosan A than toward linear beta-1,3 glucan substrates. Sometimes these two domains are found associated with other domains such as in the Catenulispora acidiphila DSM 44928 carbohydrate binding family 6 protein in which they are positioned N-terminal of a carbohydrate binding module, family 6 (CBM_6) domain. In the Cellulosimicrobium cellulans, glucan endo-1,3-beta-glucosidase, they are positioned N-terminal of a RICIN, carbohydrate-binding domain, and in the Salinispora tropica CNB-440, coagulation factor 5/8 C-terminal domain (FA58C) protein, they are positioned C-terminal of two FA58C domains which are proposed to function as cell surface-attached, carbohydrate-binding domain. This FA58C-containing protein has an internal peptide deletion (of approx. 44 residues) in the LPHase domain II. |
| pfam16483 | Glyco_hydro_64 | 6.38e-67 | 139 | 445 | 1 | 370 | Beta-1,3-glucanase. Family 64 glycoside hydrolases have beta-1,3-glucanase activity. |
| cd09216 | GH64-LPHase-like | 3.36e-34 | 149 | 443 | 6 | 350 | glycoside hydrolase family 64: laminaripentaose-producing, beta-1,3-glucanase (LPHase)-like. This subfamily is represented by the laminaripentaose-producing, beta-1,3-glucanase (LPHase) of Streptomyces matensis and related bacterial and ascomycete proteins. LPHase is a member of glycoside hydrolase family 64 (GH64), it is an inverting enzyme involved in the cleavage of long-chain polysaccharide beta-1,3-glucans, into specific pentasaccharide oligomers. LPHase is a two-domain crescent fold structure: one domain is composed of 10 beta-strands, eight coming from the N-terminus of the protein and two from the C-terminal region, and the protein has a second inserted domain; this cd includes both domains. This protein has an electronegative, substrate-binding cleft, and conserved Glu and Asp residues involved in the cleavage of the beta-1,3-glucan, laminarin, a plant and fungal cell wall component. Among bacteria, many beta-1,3-glucanases are implicated in fungal cell wall degradation. Also included in this family is GluB , the beta-1,3-glucanase B from Lysobacter enzymogenes Strain N4-7. Recombinant GluB demonstrated higher relative activity toward the branched-chain beta-1,3 glucan substrate zymosan A than toward linear beta-1,3 glucan substrates. Sometimes these two domains are found associated with other domains such as in the Catenulispora acidiphila DSM 44928 carbohydrate binding family 6 protein in which they are positioned N-terminal of a carbohydrate binding module, family 6 (CBM_6) domain. In the Cellulosimicrobium cellulans, glucan endo-1,3-beta-glucosidase, they are positioned N-terminal of a RICIN, carbohydrate-binding domain. |
| cd09220 | GH64-GluB-like | 3.26e-25 | 157 | 446 | 13 | 369 | glycoside hydrolase family 64: beta-1,3-glucanase B (GluB)-like. This subfamily is represented by GluB, beta-1,3-glucanase B , from Lysobacter enzymogenes Strain N4-7 and related bacterial and ascomycete proteins. GluB is a member of the glycoside hydrolase family 64 (GH64) involved in the cleavage of long-chain polysaccharide beta-1,3-glucans, into specific pentasaccharide oligomers. Among bacteria, many beta-1,3-glucanases are implicated in fungal cell wall degradation. GluB possesses the conserved Glu and Asp residues required to cleave substrate beta-1,3-glucans. Recombinant GluB demonstrated higher relative activity toward the branched-chain beta-1,3 glucan substrate zymosan A than toward linear beta-1,3 glucan substrates. Based on the structure of laminaripentaose-producing, beta-1,3-glucanase (LPHase) of Streptomyces matensis, which belongs to the same family as GluB but to a different subfamily, this cd is a two-domain model. Sometimes these two domains are found associated with other domains such as in the Catenulispora acidiphila DSM 44928 carbohydrate binding family 6 protein in which they are positioned N-terminal of a carbohydrate binding module, family 6 (CBM_6) domain. |
| cd08961 | GH64-TLP-SF | 2.78e-04 | 203 | 284 | 26 | 111 | glycoside hydrolase family 64 (beta-1,3-glucanases which produce specific pentasaccharide oligomers) and thaumatin-like proteins. This superfamily includes glycoside hydrolases of family 64 (GH64), these are mostly bacterial beta-1,3-glucanases which cleave long-chain polysaccharide beta-1,3-glucans, into specific pentasaccharide oligomers and are implicated in fungal cell wall degradation. Also included in this superfamily are thaumatin, the sweet-tasting protein from the African berry Thaumatococcus daniellii, and thaumatin-like proteins (TLPs) which are involved in host defense and a wide range of developmental processes in fungi, plants, and animals. Like GH64s, some TLPs also hydrolyze the beta-1,3-glucans of the type commonly found in fungal walls. Plant TLPs are classified as pathogenesis-related (PR) protein family 5 (PR5), their expression is induced by environmental stresses such as pathogen/pest attack, drought and cold. Several members of the plant TLP family have been reported as food allergens from fruits, and pollen allergens from conifers. Streptomyces matensis laminaripentaose-producing, beta-1,3-glucanase (GH64-LPHase), and TLPs have in common, a core N-terminal barrel domain (domain I) composed of 10 beta-strands, two coming from the C-terminal region of the protein. In TLPs, this core domain is flanked by two shorter domains (domains II and III). Small TLPs, such as Triticum aestivum thaumatin-like xylanase inhibitor, have a deletion in the third domain (domain II). GH64-LPHase has a second C-terminal domain which corresponds positional to, but is much larger than, domain III of TLP. GH64-LPHase and TLPs are described as crescent-fold structures. Critical functional residues, common to GH64-LPHase and TLPs are a Glu and an Asp residue. LPHase has an electronegative, substrate-binding cleft and the afore mentioned conserved Glu and Asp residues are the catalytic residues essential for beta-1,3-glucan cleavage. In TLPs, these residues are two of the four conserved residues which contribute to the strong electronegative character of the cleft which is associated with the antifungal activity of TLPs. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QAV19347.1 | 1.80e-307 | 1 | 447 | 1 | 448 |
| QOT12033.1 | 6.39e-299 | 1 | 447 | 2 | 449 |
| ACX65717.1 | 1.06e-297 | 2 | 447 | 3 | 449 |
| ANY76771.1 | 1.54e-292 | 1 | 447 | 1 | 448 |
| AWP27873.1 | 5.20e-282 | 1 | 447 | 1 | 449 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5H9X_A | 1.33e-219 | 27 | 447 | 23 | 444 | Crystalstructure of GH family 64 laminaripentaose-producing beta-1,3-glucanase from Paenibacillus barengoltzii [Paenibacillus barengoltzii] |
| 5H9Y_A | 1.09e-218 | 27 | 447 | 23 | 444 | Crystalstructure of GH family 64 laminaripentaose-producing beta-1,3-glucanase from Paenibacillus barengoltzii complexed with laminarihexaose. [Paenibacillus barengoltzii] |
| 5H4E_A | 1.24e-71 | 138 | 447 | 6 | 375 | Crystalstructure of a beta-1,3-glucanase domain (GH64) from Clostridium beijerinckii [Clostridium beijerinckii NCIMB 8052] |
| 6A6C_A | 1.37e-29 | 28 | 112 | 1 | 84 | Crystalstructure of carbohydrate-binding module family 56 beta-1,3-glucan binding domain [Paenibacillus barengoltzii] |
| 5T7A_A | 1.46e-15 | 29 | 113 | 34 | 117 | ChainA, BH0236 protein [Halalkalibacterium halodurans C-125],5T7A_B Chain B, BH0236 protein [Halalkalibacterium halodurans C-125] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P22222 | 1.21e-09 | 215 | 445 | 105 | 394 | Glucan endo-1,3-beta-glucosidase OS=Cellulosimicrobium cellulans OX=1710 PE=1 SV=1 |
| Q59146 | 1.21e-09 | 215 | 445 | 105 | 394 | Glucan endo-1,3-beta-glucosidase OS=Arthrobacter sp. (strain YCWD3) OX=79671 GN=glcI PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
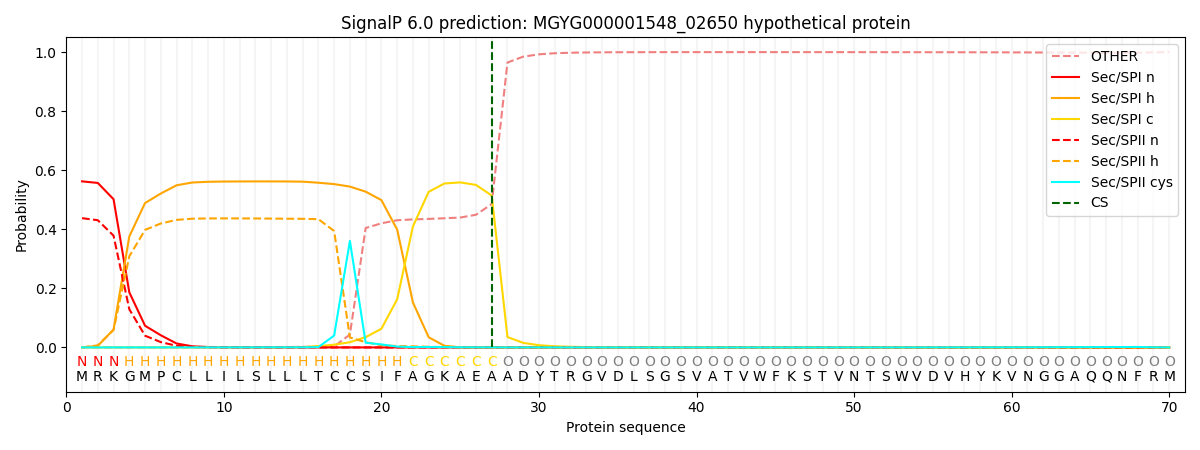
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000463 | 0.553165 | 0.445633 | 0.000282 | 0.000246 | 0.000198 |
