You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001549_00985
You are here: Home > Sequence: MGYG000001549_00985
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Gracilibacillus phocaeensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Bacillales_D; Amphibacillaceae; Gracilibacillus; Gracilibacillus phocaeensis | |||||||||||
| CAZyme ID | MGYG000001549_00985 | |||||||||||
| CAZy Family | GH126 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 187362; End: 188324 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH126 | 49 | 295 | 4.2e-47 | 0.7975077881619937 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QMT16627.1 | 7.71e-89 | 27 | 316 | 8 | 296 |
| ANU17775.1 | 2.19e-88 | 25 | 316 | 6 | 296 |
| AUD14381.1 | 6.22e-88 | 27 | 316 | 8 | 296 |
| AUD14385.1 | 1.18e-86 | 34 | 316 | 50 | 332 |
| ALS76552.1 | 1.18e-86 | 34 | 316 | 50 | 332 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6R2M_A | 8.02e-48 | 27 | 319 | 21 | 313 | Crystalstructure of PssZ from Listeria monocytogenes [Listeria monocytogenes],6R2M_B Crystal structure of PssZ from Listeria monocytogenes [Listeria monocytogenes] |
| 3REN_A | 6.56e-07 | 55 | 286 | 61 | 301 | CPF_2247,a novel alpha-amylase from Clostridium perfringens [Clostridium perfringens],3REN_B CPF_2247, a novel alpha-amylase from Clostridium perfringens [Clostridium perfringens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O31486 | 9.29e-27 | 37 | 280 | 53 | 303 | Putative lipoprotein YdaJ OS=Bacillus subtilis (strain 168) OX=224308 GN=ydaJ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
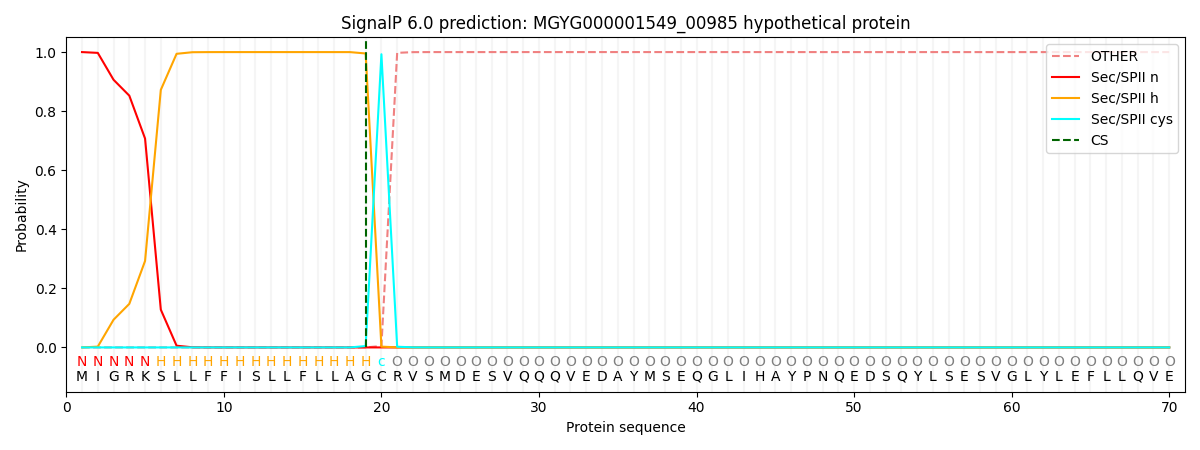
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000050 | 0.000000 | 0.000000 | 0.000000 |
