You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001549_02081
You are here: Home > Sequence: MGYG000001549_02081
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
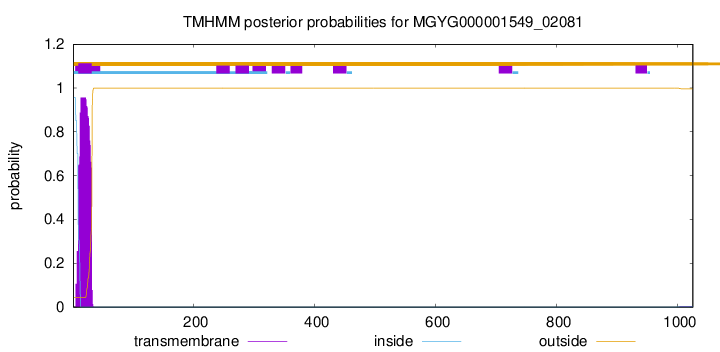
TMHMM annotations
Basic Information help
| Species | Gracilibacillus phocaeensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Bacillales_D; Amphibacillaceae; Gracilibacillus; Gracilibacillus phocaeensis | |||||||||||
| CAZyme ID | MGYG000001549_02081 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 584828; End: 587905 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4193 | LytD | 2.00e-59 | 712 | 959 | 19 | 245 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| smart00047 | LYZ2 | 8.84e-25 | 786 | 950 | 2 | 147 | Lysozyme subfamily 2. Eubacterial enzymes distantly related to eukaryotic lysozymes. |
| NF000535 | MSCRAMM_SdrC | 6.63e-17 | 140 | 337 | 714 | 899 | MSCRAMM family adhesin SdrC. Features of this protein family include a YSIRK-type signal peptide at the N-terminus and a variable-length C-terminal region of Ser-Asp (SD) repeats followed by an LPXTG motif for surface immobilization by sortase. |
| NF000535 | MSCRAMM_SdrC | 7.94e-17 | 135 | 332 | 715 | 901 | MSCRAMM family adhesin SdrC. Features of this protein family include a YSIRK-type signal peptide at the N-terminus and a variable-length C-terminal region of Ser-Asp (SD) repeats followed by an LPXTG motif for surface immobilization by sortase. |
| COG5271 | MDN1 | 4.15e-16 | 42 | 339 | 3825 | 4135 | Midasin, AAA ATPase with vWA domain, involved in ribosome maturation [Translation, ribosomal structure and biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QHS23523.1 | 3.75e-220 | 355 | 1023 | 175 | 850 |
| SQI62956.1 | 2.96e-219 | 351 | 1021 | 31 | 722 |
| QTN00811.1 | 7.68e-206 | 352 | 1021 | 229 | 912 |
| VEF49624.1 | 2.41e-191 | 341 | 1023 | 26 | 725 |
| QWC22601.1 | 4.95e-164 | 356 | 959 | 353 | 999 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6FXO_A | 1.54e-41 | 765 | 958 | 62 | 243 | ChainA, Bifunctional autolysin [Staphylococcus aureus subsp. aureus Mu50] |
| 6U0O_B | 8.39e-35 | 701 | 946 | 40 | 263 | ChainB, LYZ2 domain-containing protein [Staphylococcus aureus subsp. aureus NCTC 8325] |
| 6FXP_A | 3.30e-34 | 701 | 946 | 10 | 233 | ChainA, Uncharacterized protein [Staphylococcus aureus subsp. aureus Mu50],6FXP_B Chain B, Uncharacterized protein [Staphylococcus aureus subsp. aureus Mu50] |
| 4PI7_A | 4.22e-30 | 718 | 939 | 8 | 208 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PI9_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PIA_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
| 4PI8_A | 2.66e-29 | 718 | 939 | 8 | 208 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P39848 | 3.37e-51 | 684 | 958 | 622 | 879 | Beta-N-acetylglucosaminidase OS=Bacillus subtilis (strain 168) OX=224308 GN=lytD PE=1 SV=1 |
| O33635 | 3.80e-38 | 742 | 958 | 1130 | 1334 | Bifunctional autolysin OS=Staphylococcus epidermidis OX=1282 GN=atl PE=1 SV=1 |
| Q5HQB9 | 3.80e-38 | 742 | 958 | 1130 | 1334 | Bifunctional autolysin OS=Staphylococcus epidermidis (strain ATCC 35984 / RP62A) OX=176279 GN=atl PE=3 SV=1 |
| Q8CPQ1 | 1.99e-37 | 742 | 958 | 1130 | 1334 | Bifunctional autolysin OS=Staphylococcus epidermidis (strain ATCC 12228 / FDA PCI 1200) OX=176280 GN=atl PE=3 SV=1 |
| Q99V41 | 1.27e-36 | 765 | 958 | 1066 | 1247 | Bifunctional autolysin OS=Staphylococcus aureus (strain N315) OX=158879 GN=atl PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000598 | 0.998661 | 0.000226 | 0.000179 | 0.000166 | 0.000153 |