You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001549_04319
You are here: Home > Sequence: MGYG000001549_04319
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
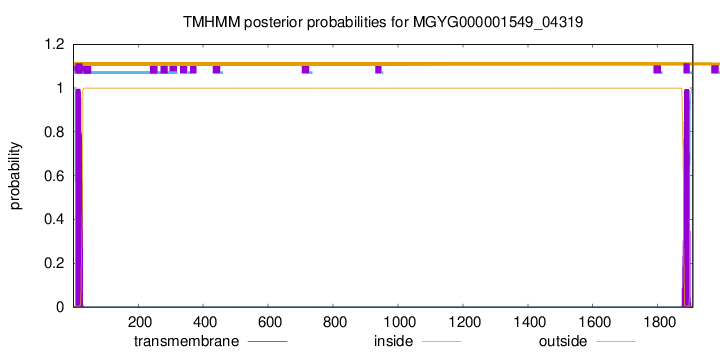
TMHMM annotations
Basic Information help
| Species | Gracilibacillus phocaeensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Bacillales_D; Amphibacillaceae; Gracilibacillus; Gracilibacillus phocaeensis | |||||||||||
| CAZyme ID | MGYG000001549_04319 | |||||||||||
| CAZy Family | GH136 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2143133; End: 2148859 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH136 | 576 | 1076 | 5.3e-117 | 0.9938900203665988 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam16403 | DUF5011 | 1.40e-23 | 1422 | 1493 | 1 | 71 | Domain of unknown function (DUF5011). This small family of proteins is functionally uncharacterized. This family is found in Bacteroides, Prevotella, and Parabateroides. Proteins in this family are around 230 amino acids in length. |
| pfam16403 | DUF5011 | 4.55e-23 | 1501 | 1572 | 1 | 71 | Domain of unknown function (DUF5011). This small family of proteins is functionally uncharacterized. This family is found in Bacteroides, Prevotella, and Parabateroides. Proteins in this family are around 230 amino acids in length. |
| pfam16403 | DUF5011 | 1.14e-13 | 1345 | 1414 | 1 | 71 | Domain of unknown function (DUF5011). This small family of proteins is functionally uncharacterized. This family is found in Bacteroides, Prevotella, and Parabateroides. Proteins in this family are around 230 amino acids in length. |
| NF033679 | DNRLRE_dom | 1.02e-10 | 318 | 472 | 1 | 143 | DNRLRE domain. The DNRLRE domain, with a length of about 160 amino acids, appears typically in large, repetitive surface proteins of bacteria and archaea, sometimes repeated several times. It occurs, notably, three times in the C-terminal region of the enzyme disaggregatase from the archaeal species Methanosarcina mazei, each time with the motif DNRLRE, for which the domain is named. Archaeal proteins within this family are described particularly well by the currently more narrowly defined Pfam model, PF06848. Note that the catalytic region of disaggregatase, in the N-terminal portion of the protein, is modeled by a different HMM, PF08480. |
| NF033679 | DNRLRE_dom | 2.43e-06 | 77 | 212 | 28 | 161 | DNRLRE domain. The DNRLRE domain, with a length of about 160 amino acids, appears typically in large, repetitive surface proteins of bacteria and archaea, sometimes repeated several times. It occurs, notably, three times in the C-terminal region of the enzyme disaggregatase from the archaeal species Methanosarcina mazei, each time with the motif DNRLRE, for which the domain is named. Archaeal proteins within this family are described particularly well by the currently more narrowly defined Pfam model, PF06848. Note that the catalytic region of disaggregatase, in the N-terminal portion of the protein, is modeled by a different HMM, PF08480. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNK57279.1 | 0.0 | 5 | 1244 | 2 | 1261 |
| AIC91360.1 | 3.16e-68 | 579 | 1083 | 87 | 683 |
| AFU70744.1 | 8.19e-68 | 579 | 1147 | 87 | 750 |
| QXN79520.1 | 4.74e-66 | 577 | 1117 | 52 | 693 |
| QNR83147.1 | 4.79e-66 | 577 | 1077 | 48 | 517 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5GQC_A | 6.76e-39 | 567 | 961 | 8 | 473 | Crystalstructure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_C Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_D Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_E Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_F Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_G Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_H Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQF_A Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, lacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQF_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, lacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQG_A Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, galacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQG_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, galacto-N-biose complex [Bifidobacterium longum subsp. longum] |
| 7V6M_A | 1.76e-37 | 572 | 989 | 4 | 482 | ChainA, Fibronectin type III domain-containing protein [Tyzzerella nexilis] |
| 7V6I_A | 2.67e-35 | 572 | 974 | 9 | 497 | ChainA, Lacto-N-biosidase [Bifidobacterium saguini DSM 23967] |
| 6KQT_A | 2.27e-34 | 579 | 952 | 247 | 662 | CrystalStructure of GH136 lacto-N-biosidase from Eubacterium ramulus - native protein [Eubacterium ramulus ATCC 29099] |
| 6KQS_A | 3.02e-34 | 579 | 952 | 247 | 662 | CrystalStructure of GH136 lacto-N-biosidase from Eubacterium ramulus - selenomethionine derivative [Eubacterium ramulus ATCC 29099] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
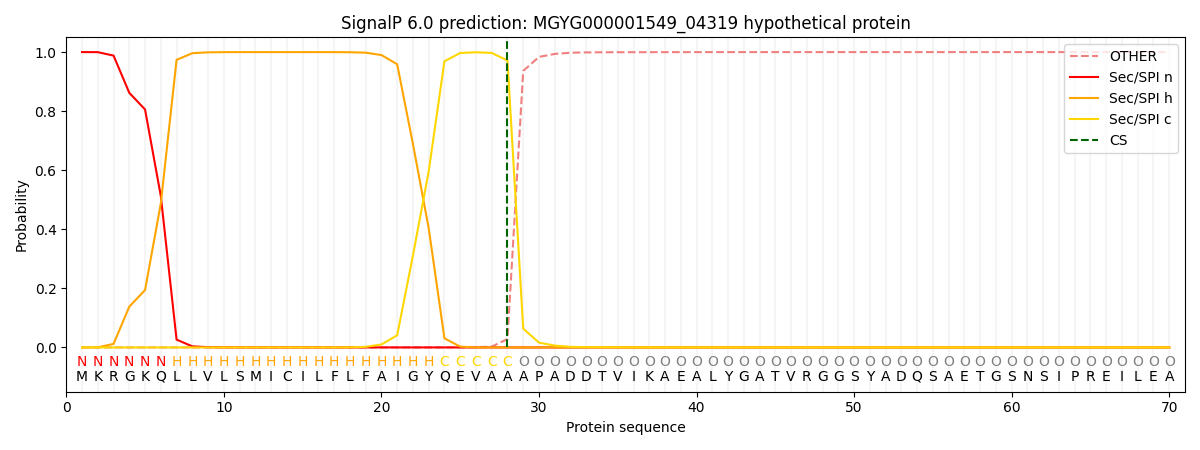
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000312 | 0.998998 | 0.000215 | 0.000173 | 0.000151 | 0.000136 |