You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001554_00359
You are here: Home > Sequence: MGYG000001554_00359
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
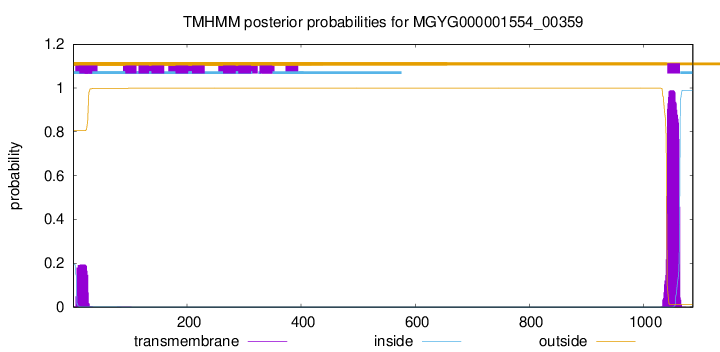
TMHMM annotations
Basic Information help
| Species | Corynebacterium phoceense | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Mycobacteriales; Mycobacteriaceae; Corynebacterium; Corynebacterium phoceense | |||||||||||
| CAZyme ID | MGYG000001554_00359 | |||||||||||
| CAZy Family | CBM5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 370236; End: 373499 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| NF033681 | ExeM_NucH_DNase | 0.0 | 244 | 825 | 1 | 545 | ExeM/NucH family extracellular endonuclease. |
| COG2374 | COG2374 | 8.29e-155 | 8 | 829 | 2 | 792 | Predicted extracellular nuclease [General function prediction only]. |
| NF033680 | exonuc_ExeM-GG | 8.59e-70 | 9 | 843 | 12 | 865 | extracellular exonuclease ExeM. ExeM, as described in Shewanella oneidensis, is a biofilm formation-associated exonuclease that cleaves extracellular DNA (eDNA), a biofilm component. Members of the ExeM family contain two or three pairs of Cys residues, presumed to form disulfide bonds, and a C-terminal GlyGly-CTERM membrane-anchoring segment. Strangely, engineered removal of the GlyGly-CTERM region did not result in net export from the cell and appearance of the enzyme in culture supernatants. |
| cd10283 | MnuA_DNase1-like | 3.33e-54 | 501 | 823 | 2 | 266 | Mycoplasma pulmonis MnuA nuclease-like. This subfamily includes Mycoplasma pulmonis MnuA, a membrane-associated nuclease related to Deoxyribonuclease 1 (DNase1 or DNase I, EC 3.1.21.1). The in vivo role of MnuA is as yet undetermined. This subfamily belongs to the large EEP (exonuclease/endonuclease/phosphatase) superfamily that contains functionally diverse enzymes that share a common catalytic mechanism of cleaving phosphodiester bonds. |
| pfam00932 | LTD | 7.47e-16 | 26 | 140 | 1 | 106 | Lamin Tail Domain. The lamin-tail domain (LTD), which has an immunoglobulin (Ig) fold, is found in Nuclear Lamins, Chlo1887 from Chloroflexus, and several bacterial proteins where it occurs with membrane associated hydrolases of the metallo-beta-lactamase,synaptojanin, and calcineurin-like phosphoesterase superfamilies. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QAY73698.1 | 3.01e-165 | 30 | 823 | 83 | 898 |
| ACZ32093.1 | 2.42e-161 | 8 | 824 | 19 | 829 |
| ANC31565.1 | 2.68e-160 | 8 | 831 | 16 | 845 |
| SDS92078.1 | 3.88e-151 | 2 | 829 | 8 | 833 |
| QAY71724.1 | 5.24e-151 | 30 | 831 | 38 | 846 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
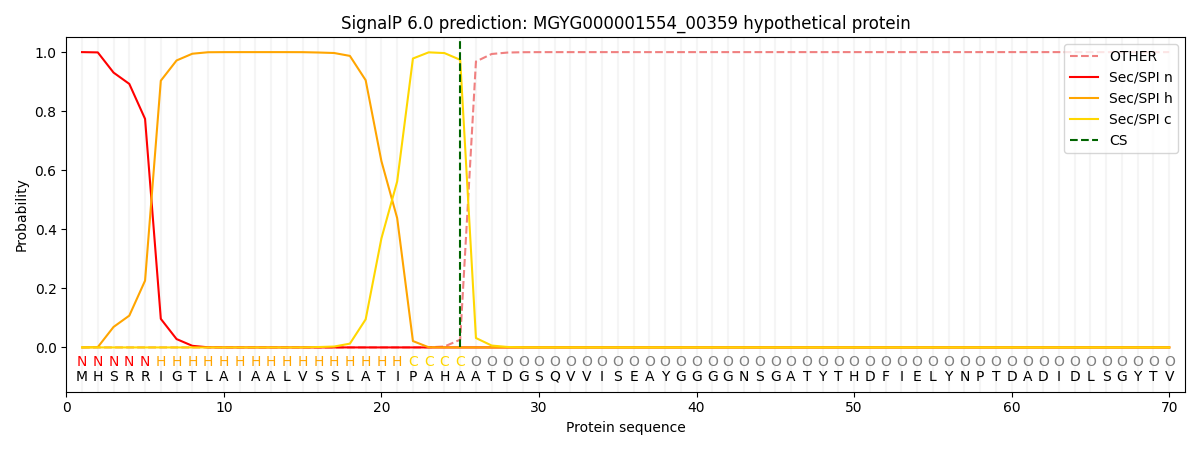
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000382 | 0.998667 | 0.000247 | 0.000279 | 0.000215 | 0.000177 |