You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001554_00460
You are here: Home > Sequence: MGYG000001554_00460
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Corynebacterium phoceense | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Mycobacteriales; Mycobacteriaceae; Corynebacterium; Corynebacterium phoceense | |||||||||||
| CAZyme ID | MGYG000001554_00460 | |||||||||||
| CAZy Family | CE1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 471226; End: 472227 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE1 | 91 | 305 | 4.9e-25 | 0.9515418502202643 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3509 | LpqC | 1.47e-24 | 91 | 332 | 41 | 308 | Poly(3-hydroxybutyrate) depolymerase [Secondary metabolites biosynthesis, transport and catabolism]. |
| COG1506 | DAP2 | 2.65e-12 | 94 | 253 | 375 | 575 | Dipeptidyl aminopeptidase/acylaminoacyl peptidase [Amino acid transport and metabolism]. |
| COG4099 | COG4099 | 7.56e-11 | 170 | 247 | 250 | 326 | Predicted peptidase [General function prediction only]. |
| TIGR01840 | esterase_phb | 5.55e-10 | 173 | 225 | 79 | 131 | esterase, PHB depolymerase family. This model describes a subfamily among lipases of the ab-hydrolase family. This subfamily includes bacterial depolymerases for poly(3-hydroxybutyrate) (PHB) and related polyhydroxyalkanoates (PHA), as well as acetyl xylan esterases, feruloyl esterases, and others from fungi. [Fatty acid and phospholipid metabolism, Degradation] |
| COG0400 | YpfH | 1.60e-08 | 168 | 247 | 74 | 157 | Predicted esterase [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CCA72326.1 | 3.38e-22 | 94 | 330 | 42 | 268 |
| QBZ61744.1 | 1.18e-21 | 68 | 329 | 21 | 267 |
| ALO15718.1 | 3.83e-21 | 94 | 330 | 32 | 288 |
| VBB80928.1 | 5.56e-20 | 94 | 329 | 41 | 266 |
| CDP29168.1 | 1.47e-19 | 94 | 329 | 41 | 266 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3WYD_A | 1.24e-09 | 99 | 247 | 23 | 165 | C-terminalesterase domain of LC-Est1 [uncultured organism],3WYD_B C-terminal esterase domain of LC-Est1 [uncultured organism] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q7RWX8 | 2.67e-16 | 65 | 330 | 23 | 288 | Feruloyl esterase D OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=faeD-3.544 PE=1 SV=1 |
| Q5B2G3 | 2.49e-15 | 97 | 279 | 43 | 211 | Feruloyl esterase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=faeC PE=1 SV=1 |
| A2QYU7 | 4.64e-15 | 97 | 300 | 43 | 236 | Probable feruloyl esterase C OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=faeC PE=3 SV=1 |
| A1CC33 | 5.70e-14 | 97 | 279 | 45 | 213 | Probable feruloyl esterase C OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=faeC-2 PE=3 SV=1 |
| B8N7Z6 | 7.57e-14 | 97 | 255 | 43 | 198 | Probable feruloyl esterase C OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=faeC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
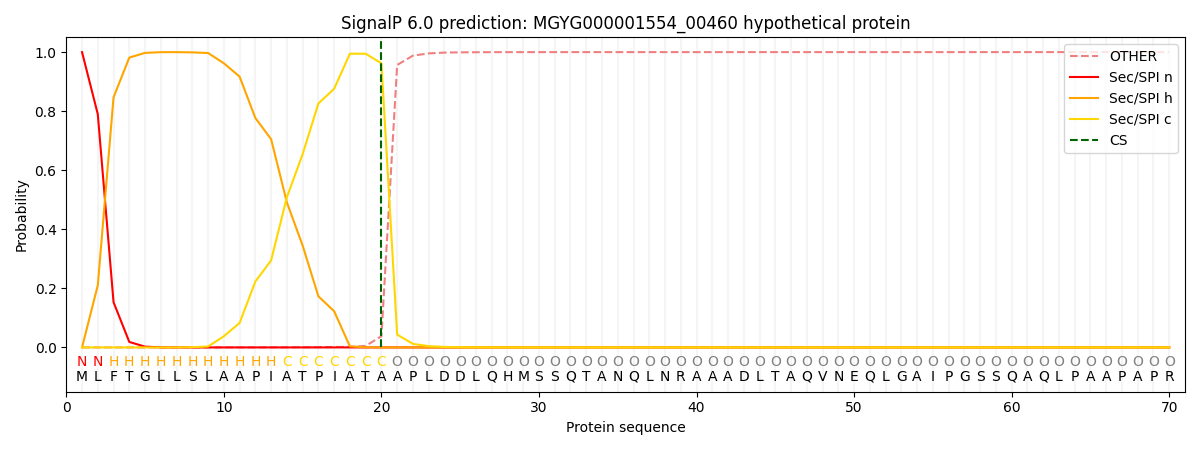
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000464 | 0.998693 | 0.000214 | 0.000220 | 0.000205 | 0.000198 |
