You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001554_02416
You are here: Home > Sequence: MGYG000001554_02416
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
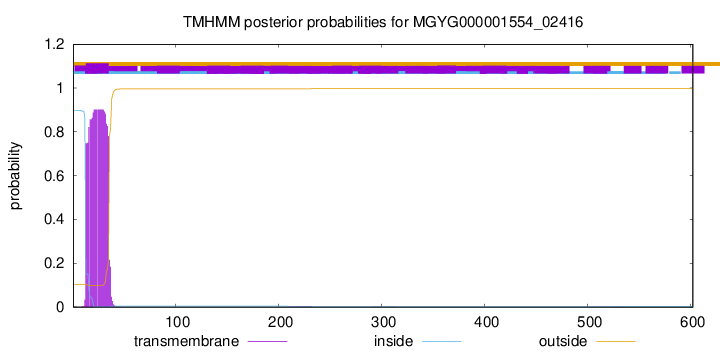
TMHMM annotations
Basic Information help
| Species | Corynebacterium phoceense | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Mycobacteriales; Mycobacteriaceae; Corynebacterium; Corynebacterium phoceense | |||||||||||
| CAZyme ID | MGYG000001554_02416 | |||||||||||
| CAZy Family | GT4 | |||||||||||
| CAZyme Description | Glycosyltransferase Gtf1 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1584666; End: 1586474 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02397 | Bac_transf | 1.36e-80 | 11 | 190 | 1 | 179 | Bacterial sugar transferase. This Pfam family represents a conserved region from a number of different bacterial sugar transferases, involved in diverse biosynthesis pathways. |
| COG2148 | WcaJ | 1.29e-72 | 3 | 195 | 33 | 224 | Sugar transferase involved in LPS biosynthesis (colanic, teichoic acid) [Cell wall/membrane/envelope biogenesis]. |
| TIGR03025 | EPS_sugtrans | 3.97e-71 | 7 | 195 | 254 | 443 | exopolysaccharide biosynthesis polyprenyl glycosylphosphotransferase. Members of this family are generally found near other genes involved in the biosynthesis of a variety of exopolysaccharides. These proteins consist of two fused domains, an N-terminal hydrophobic domain of generally low conservation and a highly conserved C-terminal sugar transferase domain (pfam02397). Characterized and partially characterized members of this subfamily include Salmonella WbaP (originally RfbP), E. coli WcaJ, Methylobacillus EpsB, Xanthomonas GumD, Vibrio CpsA, Erwinia AmsG, Group B Streptococcus CpsE (originally CpsD), and Streptococcus suis Cps2E. Each of these is believed to act in transferring the sugar from, for instance, UDP-glucose or UDP-galactose, to a lipid carrier such as undecaprenyl phosphate as the first (priming) step in the synthesis of an oligosaccharide "block". This function is encoded in the C-terminal domain. The liposaccharide is believed to be subsequently transferred through a "flippase" function from the cytoplasmic to the periplasmic face of the inner membrane by the N-terminal domain. Certain closely related transferase enzymes, such as Sinorhizobium ExoY and Lactococcus EpsD, lack the N-terminal domain and are not found by this model. |
| cd03811 | GT4_GT28_WabH-like | 3.17e-63 | 208 | 571 | 1 | 338 | family 4 and family 28 glycosyltransferases similar to Klebsiella WabH. This family is most closely related to the GT1 family of glycosyltransferases. WabH in Klebsiella pneumoniae has been shown to transfer a GlcNAc residue from UDP-GlcNAc onto the acceptor GalUA residue in the cellular outer core. |
| PRK10124 | PRK10124 | 6.20e-42 | 9 | 187 | 271 | 453 | putative UDP-glucose lipid carrier transferase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QOS67046.1 | 1.07e-62 | 207 | 555 | 2 | 359 |
| QBO59677.1 | 3.90e-61 | 187 | 563 | 2 | 364 |
| AXT59950.1 | 1.91e-58 | 207 | 562 | 2 | 346 |
| QWP59885.1 | 8.11e-53 | 204 | 579 | 2 | 360 |
| QWP62433.1 | 8.11e-53 | 204 | 579 | 2 | 360 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5W7L_A | 2.78e-36 | 5 | 202 | 5 | 196 | Structureof Campylobacter concisus PglC I57M/Q175M variant [Campylobacter concisus 13826],5W7L_B Structure of Campylobacter concisus PglC I57M/Q175M variant [Campylobacter concisus 13826] |
| 7EC2_A | 1.78e-08 | 359 | 532 | 261 | 424 | ChainA, Glycosyl transferase, group 1 family protein [Staphylococcus aureus subsp. aureus USA300],7EC2_B Chain B, Glycosyl transferase, group 1 family protein [Staphylococcus aureus subsp. aureus USA300] |
| 4X6L_A | 4.97e-07 | 350 | 542 | 235 | 436 | ChainA, TarM [Staphylococcus aureus subsp. aureus 21178],4X6L_B Chain B, TarM [Staphylococcus aureus subsp. aureus 21178],4X6L_C Chain C, TarM [Staphylococcus aureus subsp. aureus 21178],4X6L_D Chain D, TarM [Staphylococcus aureus subsp. aureus 21178],4X7P_A Chain A, TarM [Staphylococcus aureus subsp. aureus 21178],4X7P_B Chain B, TarM [Staphylococcus aureus subsp. aureus 21178] |
| 4X7M_A | 4.97e-07 | 350 | 542 | 235 | 436 | ChainA, TarM [Staphylococcus aureus subsp. aureus 21178],4X7M_B Chain B, TarM [Staphylococcus aureus subsp. aureus 21178],4X7R_A Chain A, TarM [Staphylococcus aureus subsp. aureus 21178],4X7R_B Chain B, TarM [Staphylococcus aureus subsp. aureus 21178] |
| 4WAC_A | 5.01e-07 | 350 | 542 | 240 | 441 | CrystalStructure of TarM [Staphylococcus aureus],4WAD_A Crystal Structure of TarM with UDP-GlcNAc [Staphylococcus aureus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B3FN88 | 1.28e-32 | 10 | 195 | 234 | 421 | UDP-N-acetylgalactosamine-undecaprenyl-phosphate N-acetylgalactosaminephosphotransferase OS=Aeromonas hydrophila OX=644 GN=wecA PE=1 SV=2 |
| Q0P9D0 | 2.05e-32 | 5 | 195 | 1 | 184 | Undecaprenyl phosphate N,N'-diacetylbacillosamine 1-phosphate transferase OS=Campylobacter jejuni subsp. jejuni serotype O:2 (strain ATCC 700819 / NCTC 11168) OX=192222 GN=pglC PE=1 SV=1 |
| P71241 | 3.67e-31 | 9 | 187 | 272 | 454 | UDP-glucose:undecaprenyl-phosphate glucose-1-phosphate transferase OS=Escherichia coli (strain K12) OX=83333 GN=wcaJ PE=1 SV=2 |
| P71062 | 1.82e-30 | 9 | 206 | 2 | 193 | Uncharacterized sugar transferase EpsL OS=Bacillus subtilis (strain 168) OX=224308 GN=epsL PE=2 SV=1 |
| Q44576 | 1.57e-29 | 11 | 187 | 338 | 522 | Undecaprenyl-phosphate glucose phosphotransferase OS=Komagataeibacter xylinus OX=28448 GN=aceA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
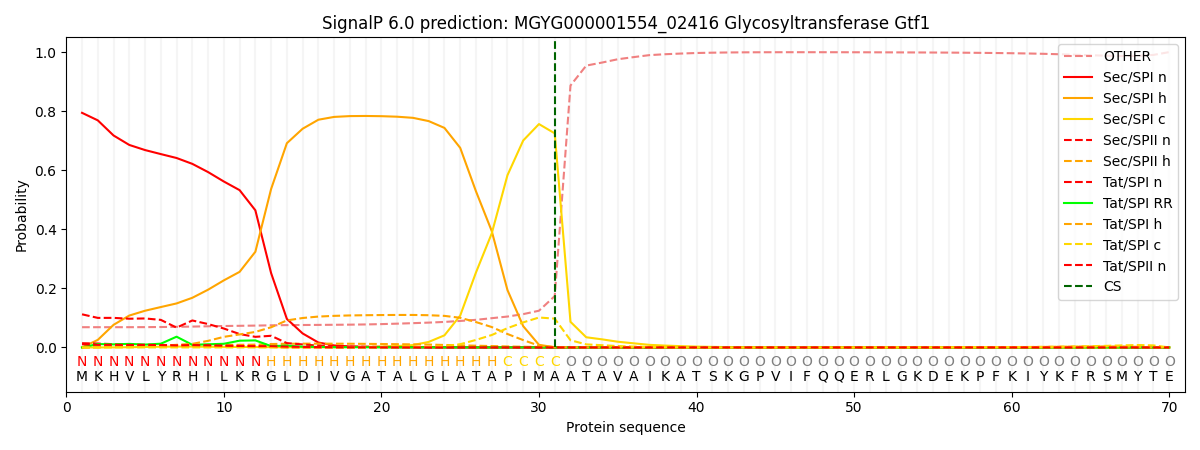
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.082357 | 0.771222 | 0.015567 | 0.117500 | 0.012516 | 0.000814 |