You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001560_01922
You are here: Home > Sequence: MGYG000001560_01922
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
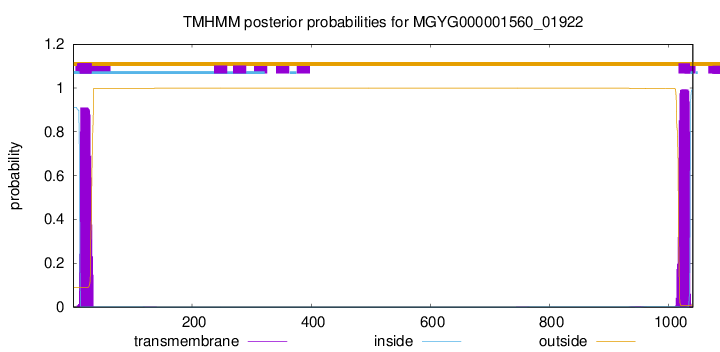
TMHMM annotations
Basic Information help
| Species | Massilioclostridium coli | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Massilioclostridium; Massilioclostridium coli | |||||||||||
| CAZyme ID | MGYG000001560_01922 | |||||||||||
| CAZy Family | GH106 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 543362; End: 546487 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH106 | 41 | 950 | 3.4e-123 | 0.9490291262135923 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17132 | Glyco_hydro_106 | 2.23e-38 | 190 | 738 | 335 | 869 | alpha-L-rhamnosidase. |
| TIGR01167 | LPXTG_anchor | 0.002 | 1011 | 1040 | 3 | 32 | LPXTG-motif cell wall anchor domain. This model describes the LPXTG motif-containing region found at the C-terminus of many surface proteins of Streptococcus and Streptomyces species. Cleavage between the Thr and Gly by sortase or a related enzyme leads to covalent anchoring at the new C-terminal Thr to the cell wall. Hits that do not lie at the C-terminus or are not found in Gram-positive bacteria are probably false-positive. A common feature of this proteins containing this domain appears to be a high proportion of charged and zwitterionic residues immediatedly upstream of the LPXTG motif. This model differs from other descriptions of the LPXTG region by including a portion of that upstream charged region. [Cell envelope, Other] |
| pfam00746 | Gram_pos_anchor | 0.005 | 1002 | 1040 | 1 | 40 | LPXTG cell wall anchor motif. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QTU60664.1 | 1.19e-130 | 37 | 976 | 41 | 951 |
| QTU52635.1 | 1.19e-130 | 37 | 976 | 41 | 951 |
| QTU44519.1 | 1.19e-130 | 37 | 976 | 41 | 951 |
| CBG74689.1 | 1.19e-130 | 37 | 976 | 41 | 951 |
| QFG20461.1 | 1.15e-129 | 42 | 976 | 49 | 976 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6Q2F_A | 2.21e-22 | 200 | 947 | 407 | 1100 | Structureof Rhamnosidase from Novosphingobium sp. PP1Y [Novosphingobium sp. PP1Y] |
| 5MQM_A | 6.37e-11 | 213 | 948 | 380 | 1060 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron],5MQN_A Glycoside hydrolase BT_0986 [Bacteroides thetaiotaomicron] |
| 5MWK_A | 1.44e-10 | 213 | 948 | 380 | 1060 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
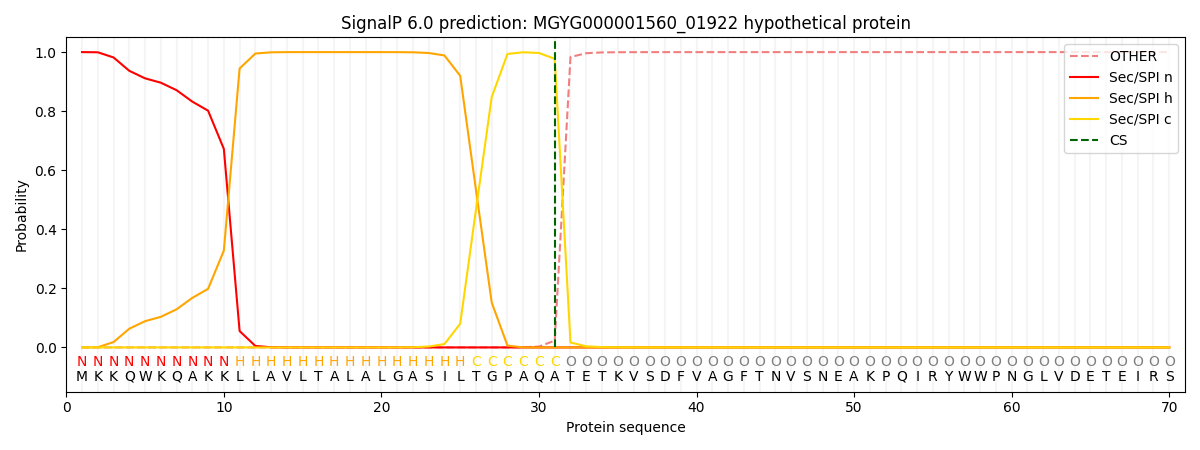
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000298 | 0.999002 | 0.000232 | 0.000164 | 0.000157 | 0.000136 |