You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001562_00155
You are here: Home > Sequence: MGYG000001562_00155
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes timonensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; Alistipes timonensis | |||||||||||
| CAZyme ID | MGYG000001562_00155 | |||||||||||
| CAZy Family | PL12 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 7317; End: 10229 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL12 | 695 | 836 | 1.6e-51 | 0.9923664122137404 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam16889 | Hepar_II_III_N | 2.22e-130 | 319 | 674 | 1 | 344 | Heparinase II/III N-terminus. This is the N-terminal domain of heparinase II/III proteins. It is a toroid-like domain. |
| pfam16889 | Hepar_II_III_N | 7.74e-78 | 55 | 320 | 1 | 251 | Heparinase II/III N-terminus. This is the N-terminal domain of heparinase II/III proteins. It is a toroid-like domain. |
| pfam07940 | Hepar_II_III | 3.87e-24 | 692 | 882 | 9 | 192 | Heparinase II/III-like protein. This family features sequences that are similar to a region of the Flavobacterium heparinum proteins heparinase II and heparinase III. The former is known to degrade heparin and heparin sulphate, whereas the latter predominantly degrades heparin sulphate. Both are secreted into the periplasmic space upon induction with heparin. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBK62838.1 | 0.0 | 1 | 970 | 1 | 964 |
| QGT73986.1 | 8.69e-163 | 311 | 970 | 42 | 702 |
| ALJ42863.1 | 6.78e-162 | 311 | 970 | 42 | 702 |
| QMW86970.1 | 6.78e-162 | 311 | 970 | 42 | 702 |
| QUT70295.1 | 6.78e-162 | 311 | 970 | 42 | 702 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4FNV_A | 2.48e-163 | 311 | 970 | 42 | 702 | CrystalStructure of Heparinase III [Bacteroides thetaiotaomicron VPI-5482] |
| 5JMD_A | 9.19e-134 | 311 | 960 | 11 | 646 | HeparinaseIII-BT4657 gene product, Methylated Lysines [Bacteroides thetaiotaomicron VPI-5482] |
| 5JMF_A | 4.01e-126 | 311 | 960 | 11 | 646 | HeparinaseIII-BT4657 gene product [Bacteroides thetaiotaomicron VPI-5482] |
| 4MMH_A | 2.16e-101 | 319 | 942 | 10 | 606 | Crystalstructure of heparan sulfate lyase HepC from Pedobacter heparinus [Pedobacter heparinus] |
| 4MMI_A | 2.16e-101 | 319 | 942 | 10 | 606 | Crystalstructure of heparan sulfate lyase HepC mutant from Pedobacter heparinus [Pedobacter heparinus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q89YR9 | 1.36e-162 | 311 | 970 | 42 | 702 | Heparin-sulfate lyase OS=Bacteroides thetaiotaomicron (strain ATCC 29148 / DSM 2079 / JCM 5827 / CCUG 10774 / NCTC 10582 / VPI-5482 / E50) OX=226186 GN=hepC PE=1 SV=1 |
| C7EXL6 | 1.79e-133 | 314 | 970 | 25 | 666 | Heparin-sulfate lyase OS=Bacteroides stercoris OX=46506 GN=hepC PE=1 SV=1 |
| Q59289 | 1.29e-100 | 309 | 942 | 24 | 630 | Heparin-sulfate lyase OS=Pedobacter heparinus (strain ATCC 13125 / DSM 2366 / CIP 104194 / JCM 7457 / NBRC 12017 / NCIMB 9290 / NRRL B-14731 / HIM 762-3) OX=485917 GN=hepC PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
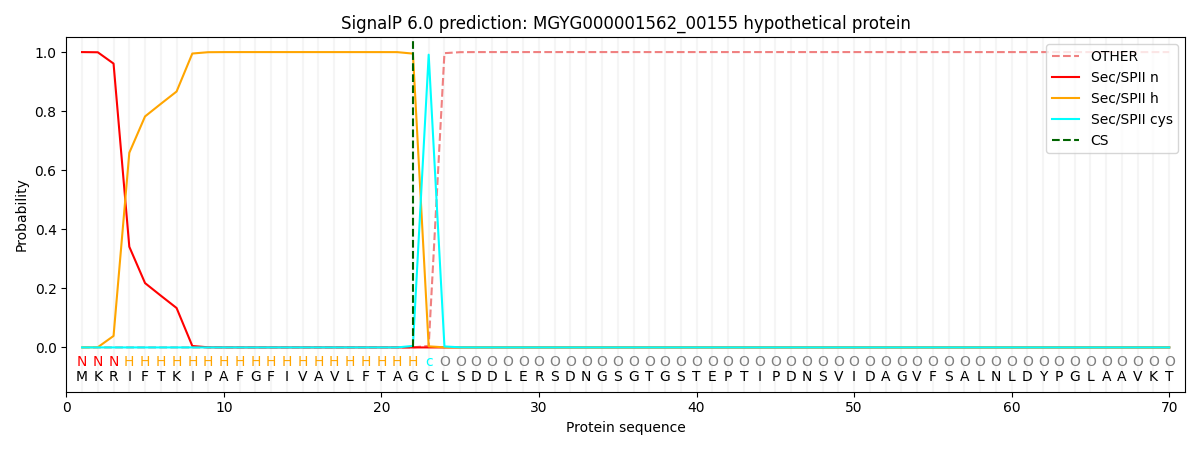
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000001 | 1.000065 | 0.000000 | 0.000000 | 0.000000 |
