You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001562_00307
You are here: Home > Sequence: MGYG000001562_00307
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes timonensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; Alistipes timonensis | |||||||||||
| CAZyme ID | MGYG000001562_00307 | |||||||||||
| CAZy Family | GH106 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 112510; End: 115719 Strand: - | |||||||||||
Full Sequence Download help
| MKNIRLCALA ACSMLLAACD GPQGLEEGFR NPSSAGVQTS VYWYWISGNV SKEGVAADLE | 60 |
| SMKRIGIDRA FIGNIGLPEA EGTPRGPVKL FTEEWYEIMH TALKTATDLG IDIGVFNCPG | 120 |
| WSQAGGPWIE PGEAMRYLAT VSAEVEGGRV VEVALPVSSP DFTDVRTVAV PLPEGVLRLD | 180 |
| ASSARVSGSA GAAKALDGDP ATALECVPGR DAVVEVKPSE TFTLRSVRIY PVHAPFAAAA | 240 |
| ELQAVKDGRT TVLRRFDIDR TNPSPEVGFD PYAPVTIACE PTEADAFRLV VSGASANGGL | 300 |
| AEVEFSALPL VERYSEKSLA KMHQTPLPYW DDYMWDRQPA GTDTTAAVAP GSVVDISDCL | 360 |
| AGDRVRWEAP AGRWMILRTG MVPTGVMNGP ALEDGAGLEV DKWSPAHLAK HYDAFVGELC | 420 |
| RRIPAADRKS WKVIVADSYE KGGQNFGDDF FEAFEARYGY DARPYLPVFS GLTVGDRDRS | 480 |
| DRFLWDLRRM MADRLSYDHI GALRELAHRD GFTTWLENYG HWGFPGEFLQ YGGQSDEVAG | 540 |
| EYWSEGELGD IENRAASSCA HIYGKNKVSA ESFTAAFSPY ARHPYRMKQR GDRFFSEGIN | 600 |
| NTLLHVYISQ AADGPKPGMN AFFGNEFNRN NTWFGQLDLF IAYLKRCNFM LQQGLNVADA | 660 |
| AYFIGEDVPK MTGVCDPALP AGYQFDYINA EVLCRDATVR DGLLTLPHGT QYRVLVLPRQ | 720 |
| ETMRPEVLER VMQLVREGAV VLGPAPTRSP SLAGYPEADG RVQAMAAELW GPETGKHVNR | 780 |
| YGKGMVFDGC SMAEVFEAIG LVPDFGHDAS LPLLFGHRTT DGAEIYFVSN QSEEPIGFTA | 840 |
| SFRVADRRPE WWQPVTGAVR DLPAWRAEGA VTEIPMRLEP LESGFVVFRK PAAESAAEFT | 900 |
| ADANFPRPEP IAAVDGAWQV RFEAPDGVGN FTLATDTLGD WSQSADERLR GFSGTATYST | 960 |
| TFEKPAADGG RILLDLNRVG VMAKVRVNGA DAGGVWTPPY RVDVTDLLRE GGNELEIEVV | 1020 |
| NTWVNRLVGD SRLPEAERRT WTPYNPYTPE SPLQESGLIG PVTLQRVAL | 1069 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH106 | 349 | 1065 | 2.6e-225 | 0.8847087378640777 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17132 | Glyco_hydro_106 | 0.0 | 32 | 862 | 4 | 869 | alpha-L-rhamnosidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBL11634.1 | 0.0 | 25 | 1067 | 32 | 1077 |
| BBL08842.1 | 0.0 | 25 | 1067 | 32 | 1077 |
| BBL00916.1 | 0.0 | 25 | 1067 | 32 | 1077 |
| QRQ58444.1 | 0.0 | 1 | 1067 | 1 | 1078 |
| ALJ46926.1 | 0.0 | 1 | 1067 | 1 | 1078 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6Q2F_A | 2.97e-152 | 25 | 1064 | 42 | 1138 | Structureof Rhamnosidase from Novosphingobium sp. PP1Y [Novosphingobium sp. PP1Y] |
| 5MQM_A | 1.18e-115 | 25 | 1062 | 31 | 1096 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron],5MQN_A Glycoside hydrolase BT_0986 [Bacteroides thetaiotaomicron] |
| 5MWK_A | 3.13e-115 | 25 | 1062 | 31 | 1096 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron] |
| 6MVF_A | 9.19e-06 | 911 | 1021 | 27 | 140 | Crystalstructure of FMN-binding beta-glucuronidase from Facaelibacterium prausnitzii L2-6 [Faecalibacterium prausnitzii L2-6],6MVF_B Crystal structure of FMN-binding beta-glucuronidase from Facaelibacterium prausnitzii L2-6 [Faecalibacterium prausnitzii L2-6],6MVF_C Crystal structure of FMN-binding beta-glucuronidase from Facaelibacterium prausnitzii L2-6 [Faecalibacterium prausnitzii L2-6],6MVF_D Crystal structure of FMN-binding beta-glucuronidase from Facaelibacterium prausnitzii L2-6 [Faecalibacterium prausnitzii L2-6],6MVF_E Crystal structure of FMN-binding beta-glucuronidase from Facaelibacterium prausnitzii L2-6 [Faecalibacterium prausnitzii L2-6],6MVF_F Crystal structure of FMN-binding beta-glucuronidase from Facaelibacterium prausnitzii L2-6 [Faecalibacterium prausnitzii L2-6] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KNA8 | 8.48e-112 | 25 | 1064 | 30 | 1155 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22040 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
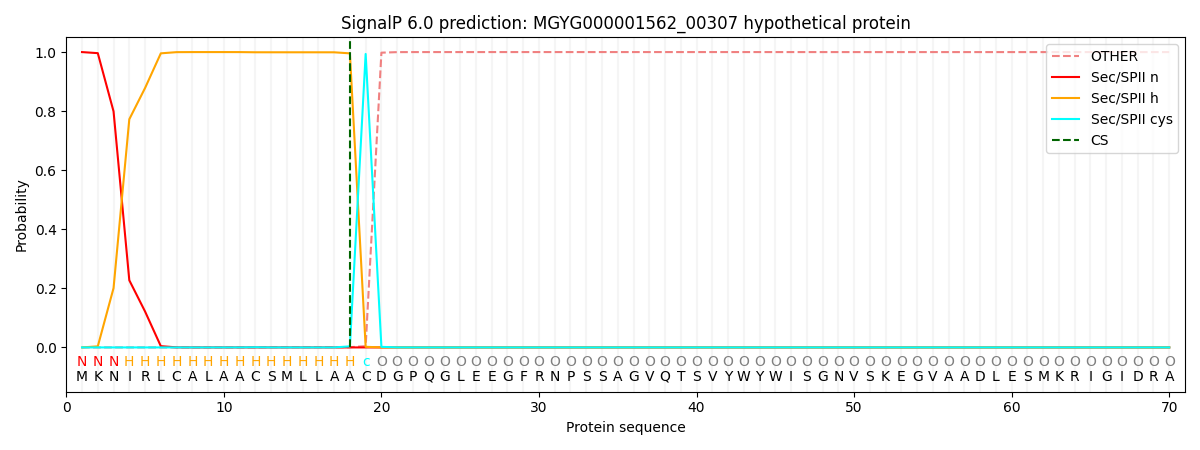
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000069 | 0.000000 | 0.000000 | 0.000000 |
