You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001562_01116
You are here: Home > Sequence: MGYG000001562_01116
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes timonensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; Alistipes timonensis | |||||||||||
| CAZyme ID | MGYG000001562_01116 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 243293; End: 246274 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 23 | 505 | 2.8e-47 | 0.5558510638297872 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 3.40e-21 | 63 | 484 | 66 | 508 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10150 | PRK10150 | 3.93e-16 | 29 | 349 | 15 | 352 | beta-D-glucuronidase; Provisional |
| PRK10340 | ebgA | 2.73e-13 | 26 | 352 | 92 | 397 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| PRK09525 | lacZ | 1.62e-09 | 139 | 374 | 194 | 434 | beta-galactosidase. |
| pfam02836 | Glyco_hydro_2_C | 4.57e-06 | 270 | 360 | 2 | 87 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADY51369.1 | 0.0 | 29 | 983 | 31 | 994 |
| QNN44806.1 | 0.0 | 1 | 978 | 5 | 992 |
| QEC78585.1 | 0.0 | 27 | 983 | 33 | 996 |
| BCI63942.1 | 0.0 | 4 | 982 | 2 | 981 |
| QJC53152.1 | 1.97e-19 | 19 | 456 | 13 | 509 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1BHG_A | 8.36e-12 | 26 | 349 | 41 | 383 | HumanBeta-Glucuronidase At 2.6 A Resolution [Homo sapiens],1BHG_B Human Beta-Glucuronidase At 2.6 A Resolution [Homo sapiens],3HN3_A Human beta-glucuronidase at 1.7 A resolution [Homo sapiens],3HN3_B Human beta-glucuronidase at 1.7 A resolution [Homo sapiens],3HN3_D Human beta-glucuronidase at 1.7 A resolution [Homo sapiens],3HN3_E Human beta-glucuronidase at 1.7 A resolution [Homo sapiens] |
| 5EUV_A | 3.93e-09 | 74 | 385 | 63 | 358 | ChainA, Beta-D-galactosidase [Paracoccus sp. 32d],5EUV_B Chain B, Beta-D-galactosidase [Paracoccus sp. 32d],5LDR_B Chain B, Beta-D-galactosidase [Paracoccus sp. 32d] |
| 5LDR_A | 3.94e-09 | 74 | 385 | 64 | 359 | ChainA, Beta-D-galactosidase [Paracoccus sp. 32d] |
| 5UJ6_A | 4.27e-09 | 57 | 391 | 73 | 418 | CrystalStructure of Bacteroides Uniformis beta-glucuronidase [Bacteroides uniformis str. 3978 T3 ii],5UJ6_B Crystal Structure of Bacteroides Uniformis beta-glucuronidase [Bacteroides uniformis str. 3978 T3 ii],6NZG_A Bacteroides uniformis beta-glucuronidase 2 covalently bound to cyclophellitol-6-carboxylate aziridine [Bacteroides uniformis],6NZG_B Bacteroides uniformis beta-glucuronidase 2 covalently bound to cyclophellitol-6-carboxylate aziridine [Bacteroides uniformis] |
| 6D50_A | 4.29e-09 | 57 | 391 | 81 | 426 | Bacteroidesuniforms beta-glucuronidase 2 bound to D-glucaro-1,5-lactone [Bacteroides uniformis str. 3978 T3 ii],6D50_B Bacteroides uniforms beta-glucuronidase 2 bound to D-glucaro-1,5-lactone [Bacteroides uniformis str. 3978 T3 ii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q4FAT7 | 2.59e-13 | 60 | 349 | 98 | 403 | Beta-glucuronidase OS=Sus scrofa OX=9823 GN=GUSB PE=3 SV=1 |
| O77695 | 3.40e-13 | 56 | 349 | 90 | 400 | Beta-glucuronidase (Fragment) OS=Chlorocebus aethiops OX=9534 GN=GUSB PE=2 SV=1 |
| Q47077 | 1.81e-12 | 139 | 392 | 195 | 464 | Beta-galactosidase OS=Enterobacter cloacae OX=550 GN=lacZ PE=3 SV=1 |
| P06760 | 3.07e-12 | 42 | 349 | 71 | 399 | Beta-glucuronidase OS=Rattus norvegicus OX=10116 GN=Gusb PE=1 SV=1 |
| Q2XQU3 | 3.12e-12 | 139 | 392 | 195 | 464 | Beta-galactosidase 2 OS=Enterobacter cloacae OX=550 GN=lacZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
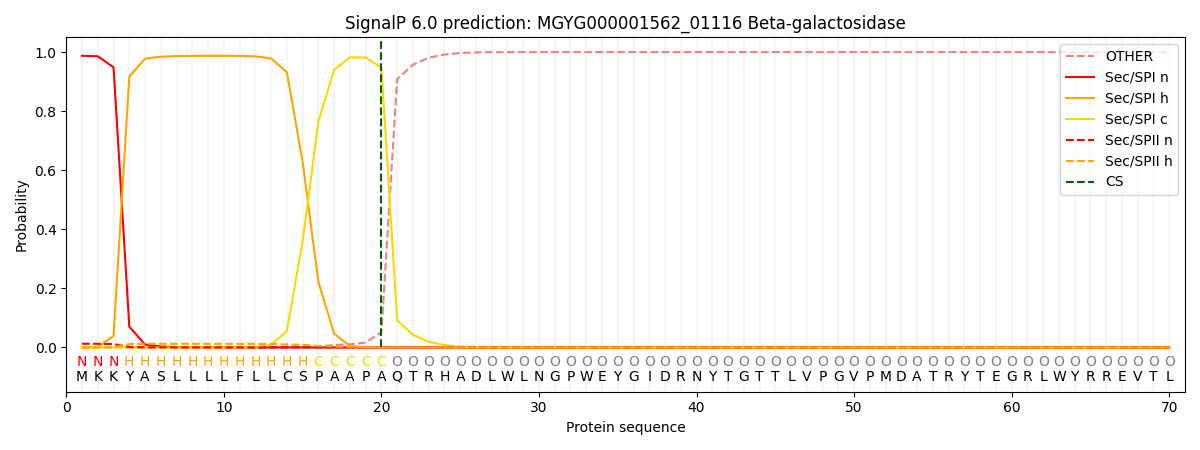
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001356 | 0.983089 | 0.014578 | 0.000347 | 0.000319 | 0.000290 |
