You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001567_00700
You are here: Home > Sequence: MGYG000001567_00700
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Coriobacteriia; Coriobacteriales; Coriobacteriaceae; Collinsella; | |||||||||||
| CAZyme ID | MGYG000001567_00700 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 79279; End: 80505 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH28 | 34 | 240 | 2.6e-29 | 0.5661538461538461 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5434 | Pgu1 | 2.35e-24 | 21 | 238 | 99 | 349 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| pfam00295 | Glyco_hydro_28 | 7.73e-08 | 146 | 226 | 88 | 176 | Glycosyl hydrolases family 28. Glycosyl hydrolase family 28 includes polygalacturonase EC:3.2.1.15 as well as rhamnogalacturonase A(RGase A), EC:3.2.1.-. These enzymes are important in cell wall metabolism. |
| PLN03010 | PLN03010 | 9.63e-07 | 116 | 226 | 133 | 248 | polygalacturonase |
| pfam13229 | Beta_helix | 2.44e-05 | 145 | 249 | 2 | 110 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| PLN02155 | PLN02155 | 0.006 | 171 | 214 | 173 | 221 | polygalacturonase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUC03291.1 | 8.12e-193 | 6 | 407 | 3 | 402 |
| AEB07770.1 | 1.22e-185 | 1 | 406 | 1 | 406 |
| AUJ85227.1 | 7.84e-97 | 21 | 403 | 24 | 400 |
| QED59087.1 | 1.48e-89 | 24 | 339 | 28 | 343 |
| ASG79845.1 | 5.51e-88 | 21 | 385 | 23 | 381 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5OLP_A | 1.97e-09 | 21 | 250 | 61 | 301 | Galacturonidase[Bacteroides thetaiotaomicron VPI-5482],5OLP_B Galacturonidase [Bacteroides thetaiotaomicron VPI-5482] |
| 2UVE_A | 8.30e-06 | 21 | 213 | 173 | 405 | Structureof Yersinia enterocolitica Family 28 Exopolygalacturonase [Yersinia enterocolitica],2UVE_B Structure of Yersinia enterocolitica Family 28 Exopolygalacturonase [Yersinia enterocolitica],2UVF_A Structure of Yersinia enterocolitica Family 28 Exopolygalacturonase in Complex with Digalaturonic Acid [Yersinia enterocolitica],2UVF_B Structure of Yersinia enterocolitica Family 28 Exopolygalacturonase in Complex with Digalaturonic Acid [Yersinia enterocolitica] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q40312 | 1.73e-07 | 29 | 224 | 48 | 234 | Polygalacturonase OS=Medicago sativa OX=3879 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
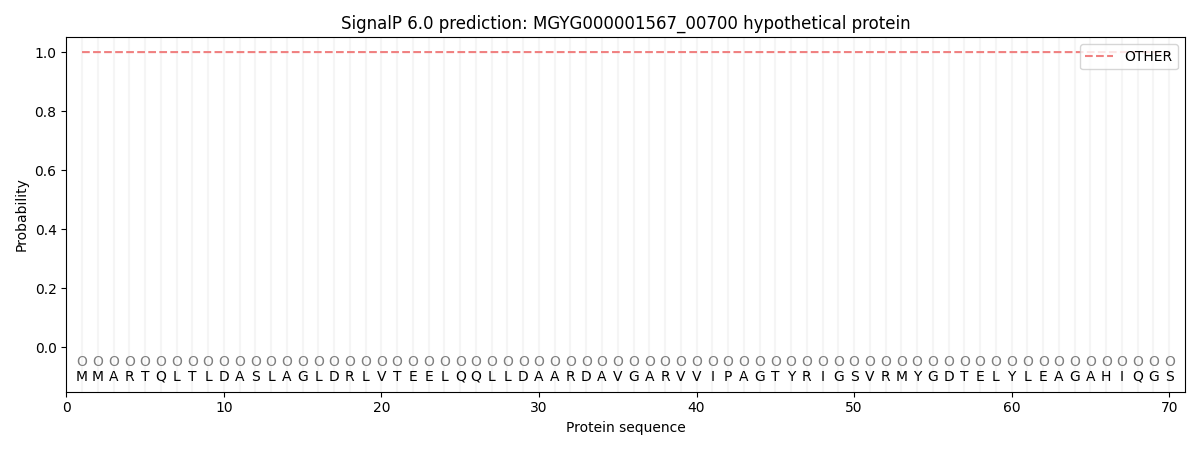
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000061 | 0.000001 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
