You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001571_00124
You are here: Home > Sequence: MGYG000001571_00124
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Lacrimispora sp000526575 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Lacrimispora; Lacrimispora sp000526575 | |||||||||||
| CAZyme ID | MGYG000001571_00124 | |||||||||||
| CAZy Family | GT84 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 125925; End: 131729 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT84 | 1216 | 1428 | 8.4e-87 | 0.986046511627907 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd11753 | GH94N_ChvB_NdvB_2_like | 1.30e-101 | 1421 | 1740 | 1 | 322 | Second GH94N domain of cyclic beta 1-2 glucan synthetase and similar domains. The glycoside hydrolase family 94 (previously known as glycosyltransferase family 36) includes cyclic beta 1-2 glucan synthetase (EC:2.4.1.20) or ChvB (encoded by the chromosomal chvB virulence gene). This second of two tandemly repeated GH94-N-terminal-like domains has not been characterized functionally. Some beta 1-2 glucan synthetases are annotated as NdvB (nodule development B) gene products, glycosyltransferases required for the synthesis of cyclic beta-(1,2)-glucans, which play a role in interactions between bacteria and plants. |
| pfam06165 | Glyco_transf_36 | 6.36e-73 | 1480 | 1727 | 2 | 247 | Glycosyltransferase family 36. The glycosyltransferase family 36 includes cellobiose phosphorylase (EC:2.4.1.20), cellodextrin phosphorylase (EC:2.4.1.49), chitobiose phosphorylase (EC:2.4.1.-). Many members of this family contain two copies of this domain. |
| cd11751 | GH94N_like_4 | 8.27e-31 | 1520 | 1734 | 7 | 223 | Glycoside hydrolase family 94 N-terminal-like domain of uncharacterized function. The glycoside hydrolase family 94 (previously known as glycosyltransferase family 36) includes cellobiose phosphorylase (EC:2.4.1.20), cellodextrin phosphorylase (EC:2.4.1.49), chitobiose phosphorylase (EC:2.4.1.-), amongst other members. Their N-terminal domain is involved in oligomerization and may play a role in catalysis, but it is separate from the catalytic domain [an (alpha/alpha)(6) barrel]. The GH64N domain, as represented by this model, is found near the N-terminus of GH94 members and related proteins with uncharacterized specificities. |
| pfam10091 | Glycoamylase | 1.15e-27 | 1217 | 1427 | 3 | 213 | Putative glucoamylase. The structure of UniProt:Q5LIB7 has an alpha/alpha toroid fold and is similar structurally to a number of glucoamylases. Most of these structural homologs are glucoamylases, involved in breaking down complex sugars (e.g. starch). The biologically relevant state is likely to be monomeric. The putative active site is located at the centre of the toroid with a well defined large cavity. |
| cd11755 | GH94N_ChBP_like | 1.38e-21 | 1473 | 1754 | 15 | 300 | N-terminal domain of chitobiose phosphorylase (ChBP) and similar proteins. The glycoside hydrolase family 94 (previously known as glycosyltransferase family 36) includes chitobiose phosphorylase (EC:2.4.1.-). This N-terminal domain is involved in oligomerization and may play a role in catalysis, but it is separate from the catalytic domain [an (alpha/alpha)(6) barrel]. Chitobiose phosphorylase catalyzes the reversible phosphate dependent hydrolysis of chitobiose [(GlcNAc)2] into alpha-GlcNAc-1-phosphate and GlcNAc. In some organisms, ChBP may be involved in the production of GlcNac-6-phosphate in intracellular pathways. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRV21370.1 | 0.0 | 4 | 1934 | 4 | 2039 |
| SET84643.1 | 0.0 | 1 | 1934 | 1 | 2041 |
| CAB1254454.1 | 0.0 | 1 | 1934 | 1 | 2038 |
| BCJ98660.1 | 0.0 | 1 | 1929 | 1 | 2035 |
| VDN47041.1 | 0.0 | 35 | 1933 | 36 | 2031 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P20471 | 5.85e-260 | 96 | 1917 | 67 | 1999 | Cyclic beta-(1,2)-glucan synthase NdvB OS=Rhizobium meliloti (strain 1021) OX=266834 GN=ndvB PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
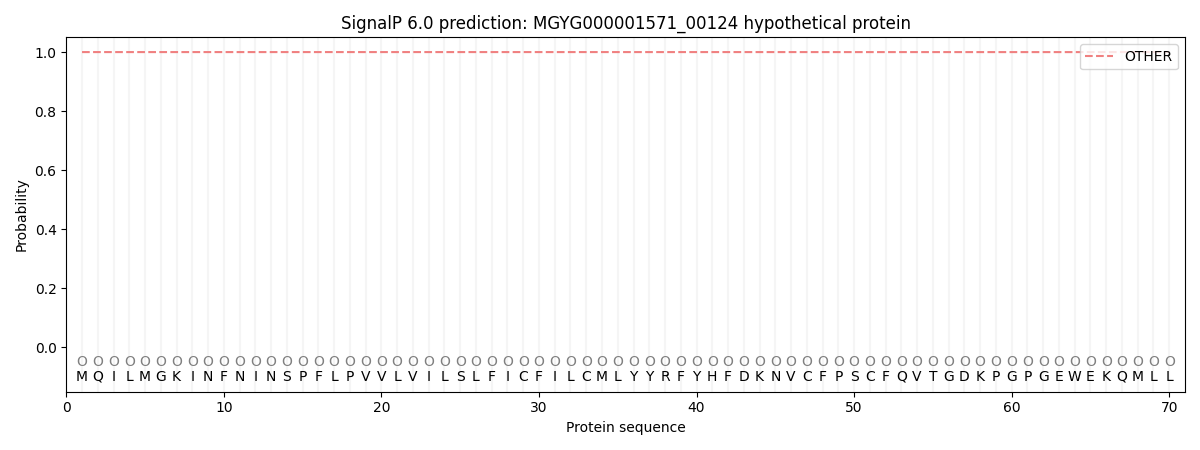
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000062 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
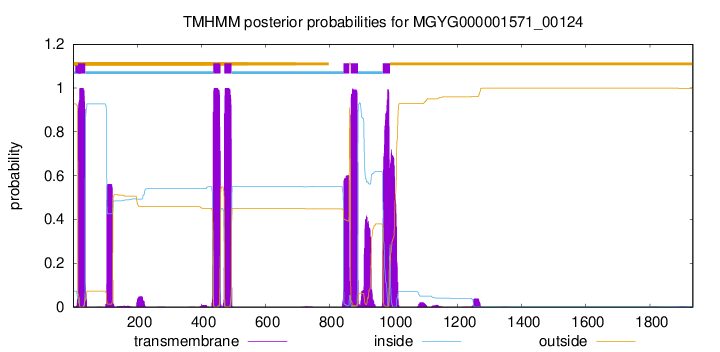
| start | end |
|---|---|
| 15 | 37 |
| 437 | 459 |
| 472 | 494 |
| 844 | 861 |
| 866 | 888 |
| 966 | 988 |
