You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001571_01470
You are here: Home > Sequence: MGYG000001571_01470
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
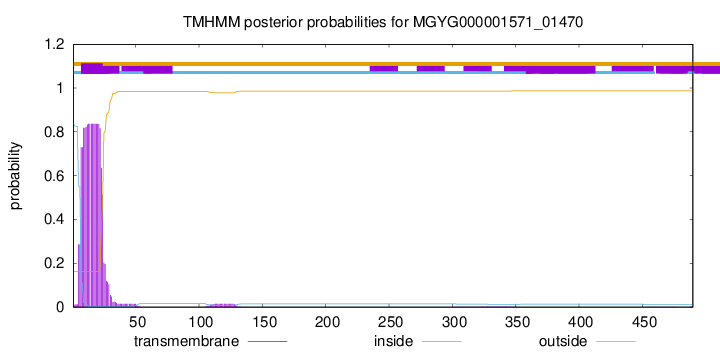
TMHMM annotations
Basic Information help
| Species | Lacrimispora sp000526575 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Lacrimispora; Lacrimispora sp000526575 | |||||||||||
| CAZyme ID | MGYG000001571_01470 | |||||||||||
| CAZy Family | GH93 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 127195; End: 128667 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH93 | 44 | 326 | 1.6e-85 | 0.9706840390879479 |
| CBM13 | 356 | 488 | 2.2e-23 | 0.6914893617021277 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam14200 | RicinB_lectin_2 | 1.29e-20 | 391 | 475 | 1 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| pfam00652 | Ricin_B_lectin | 2.93e-16 | 357 | 485 | 1 | 126 | Ricin-type beta-trefoil lectin domain. |
| pfam14200 | RicinB_lectin_2 | 9.19e-13 | 354 | 430 | 10 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| cd00161 | RICIN | 4.41e-12 | 357 | 487 | 1 | 124 | Ricin-type beta-trefoil; Carbohydrate-binding domain formed from presumed gene triplication. The domain is found in a variety of molecules serving diverse functions such as enzymatic activity, inhibitory toxicity and signal transduction. Highly specific ligand binding occurs on exposed surfaces of the compact domain sturcture. |
| pfam14200 | RicinB_lectin_2 | 1.24e-11 | 436 | 487 | 1 | 54 | Ricin-type beta-trefoil lectin domain-like. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGQ95143.1 | 2.81e-119 | 3 | 405 | 15 | 414 |
| AWV33771.1 | 4.29e-107 | 26 | 404 | 30 | 464 |
| QDY06014.1 | 3.11e-106 | 10 | 400 | 23 | 443 |
| QYX75853.1 | 3.75e-103 | 13 | 404 | 28 | 450 |
| AVV45913.1 | 7.19e-99 | 34 | 404 | 53 | 449 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2W5N_A | 1.05e-68 | 24 | 335 | 12 | 343 | ChainA, Alpha-l-arabinofuranosidase [Fusarium graminearum] |
| 2YDT_A | 5.75e-68 | 24 | 335 | 12 | 343 | ChainA, Exo-1,5-alpha-l-arabinofuranobiosidase [Fusarium graminearum],5M1Z_A Chain A, Exo-1,5-alpha-L-arabinofuranobiosidase [Fusarium graminearum] |
| 2YDP_A | 4.43e-67 | 24 | 335 | 12 | 343 | ChainA, Exo-1,5-alpha-l-arabinofuranobiosidase [Fusarium graminearum],2YDP_B Chain B, Exo-1,5-alpha-l-arabinofuranobiosidase [Fusarium graminearum],2YDP_C Chain C, Exo-1,5-alpha-l-arabinofuranobiosidase [Fusarium graminearum] |
| 2W5O_A | 1.72e-66 | 24 | 335 | 12 | 343 | ChainA, ALPHA-L-ARABINOFURANOSIDASE [Fusarium graminearum] |
| 3A71_A | 1.02e-64 | 34 | 332 | 11 | 331 | Highresolution structure of Penicillium chrysogenum alpha-L-arabinanase [Penicillium chrysogenum],3A72_A High resolution structure of Penicillium chrysogenum alpha-L-arabinanase complexed with arabinobiose [Penicillium chrysogenum] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
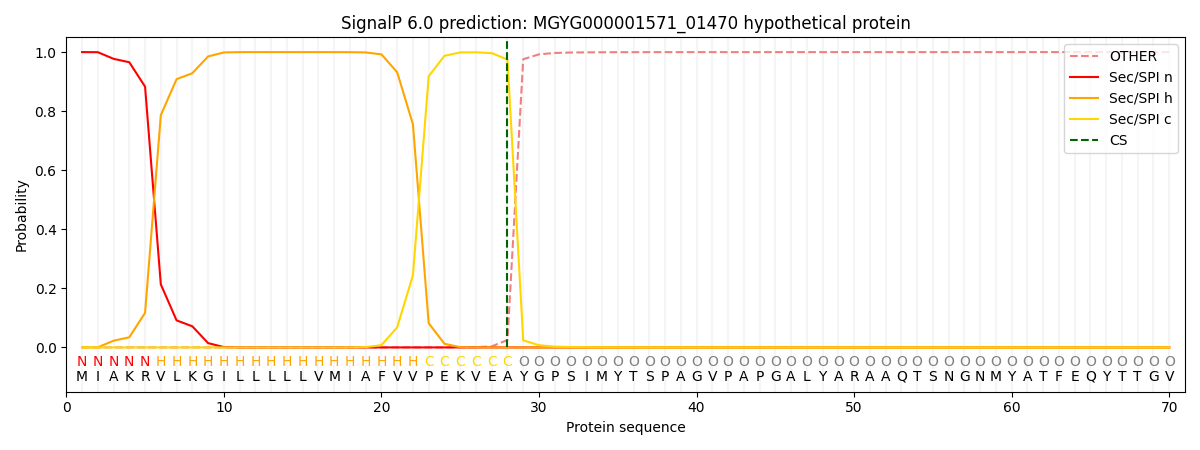
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000228 | 0.999113 | 0.000182 | 0.000157 | 0.000147 | 0.000140 |