You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001582_00186
You are here: Home > Sequence: MGYG000001582_00186
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
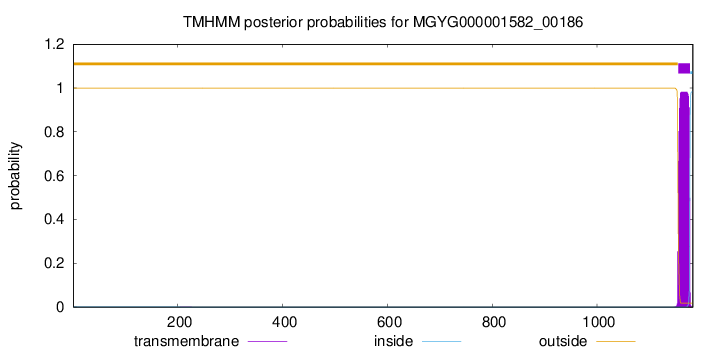
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Coriobacteriia; Coriobacteriales; Coriobacteriaceae; Collinsella; | |||||||||||
| CAZyme ID | MGYG000001582_00186 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 35; End: 3586 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 109 | 224 | 7.1e-16 | 0.8709677419354839 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd08759 | Type_III_cohesin_like | 2.53e-37 | 311 | 478 | 1 | 167 | Cohesin domain, interaction partner of dockerin. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. Two specific calcium-dependent interactions between cohesin and dockerin appear to be essential for cellulosome assembly, type I and type II. This subfamily represents type III cohesins and closely related domains. |
| pfam00754 | F5_F8_type_C | 4.46e-18 | 101 | 217 | 1 | 118 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd00057 | FA58C | 7.63e-10 | 103 | 217 | 18 | 134 | Substituted updates: Jan 31, 2002 |
| pfam00754 | F5_F8_type_C | 1.14e-09 | 807 | 944 | 5 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam07554 | FIVAR | 3.47e-08 | 960 | 1025 | 3 | 69 | FIVAR domain. This domain is found in a wide variety of contexts, but mostly occurring in cell wall associated proteins. A lack of conserved catalytic residues suggests that it is a binding domain. From context, possible substrates are hyaluronate or fibronectin (personal obs: C Yeats). This is further evidenced by. Possibly the exact substrate is N-acetyl glucosamine. Finding it in the same protein as pfam05089 further supports this proposal. It is found in the C-terminal part of Bacillus sp. Gellan lyase, which is removed during maturation. Some of the proteins it is found in are involved in methicillin resistance. The name FIVAR derives from Found In Various Architectures. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QWT17625.1 | 0.0 | 6 | 1107 | 977 | 2112 |
| QNM10857.1 | 4.14e-253 | 4 | 1105 | 934 | 2018 |
| BCT46261.1 | 2.50e-213 | 11 | 1100 | 935 | 2041 |
| QOY60737.1 | 1.18e-172 | 6 | 483 | 366 | 848 |
| BBK61154.1 | 1.46e-162 | 1 | 1106 | 957 | 2075 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4LPL_A | 9.89e-22 | 73 | 228 | 23 | 182 | Structureof CBM32-1 from a family 31 glycoside hydrolase from Clostridium perfringens [Clostridium perfringens ATCC 13124] |
| 2V72_A | 3.37e-15 | 96 | 228 | 11 | 143 | Thestructure of the family 32 CBM from C. perfringens NanJ in complex with galactose [Clostridium perfringens] |
| 4LKS_A | 2.86e-14 | 807 | 948 | 32 | 166 | Structureof CBM32-3 from a family 31 glycoside hydrolase from Clostridium perfringens in complex with galactose [Clostridium perfringens ATCC 13124],4LKS_C Structure of CBM32-3 from a family 31 glycoside hydrolase from Clostridium perfringens in complex with galactose [Clostridium perfringens ATCC 13124],4LQR_A Structure of CBM32-3 from a family 31 glycoside hydrolase from Clostridium perfringens [Clostridium perfringens ATCC 13124],4P5Y_A Structure of CBM32-3 from a family 31 glycoside hydrolase from Clostridium perfringens in complex with N-acetylgalactosamine [Clostridium perfringens ATCC 13124] |
| 6M76_A | 2.10e-12 | 5 | 94 | 874 | 963 | GH31alpha-N-acetylgalactosaminidase from Enterococcus faecalis [Enterococcus faecalis ATCC 10100],6M77_A GH31 alpha-N-acetylgalactosaminidase from Enterococcus faecalis in complex with N-acetylgalactosamine [Enterococcus faecalis ATCC 10100] |
| 7F7Q_A | 2.10e-12 | 5 | 94 | 874 | 963 | ChainA, GH31 alpha-N-acetylgalactosaminidase [Enterococcus faecalis ATCC 10100] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| E8MGH9 | 1.81e-07 | 902 | 1100 | 1610 | 1805 | Beta-L-arabinobiosidase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=hypBA2 PE=1 SV=1 |
| Q0TR53 | 1.05e-06 | 140 | 227 | 672 | 766 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain ATCC 13124 / DSM 756 / JCM 1290 / NCIMB 6125 / NCTC 8237 / Type A) OX=195103 GN=nagJ PE=1 SV=1 |
| Q8XL08 | 1.05e-06 | 140 | 227 | 672 | 766 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagJ PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
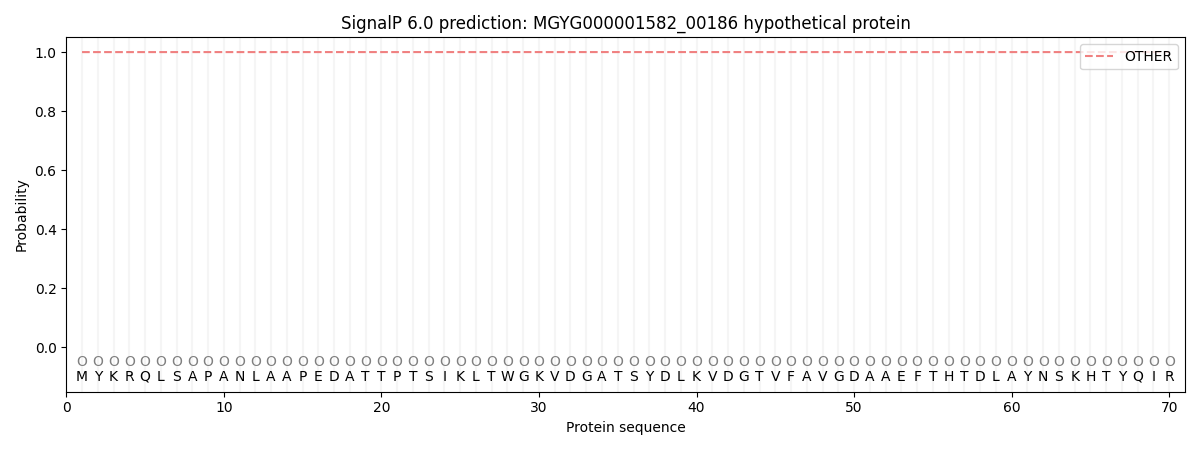
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000059 | 0.000002 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |