You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001588_00881
Basic Information
help
| Species |
|
| Lineage |
Bacteria; Actinobacteriota; Coriobacteriia; Coriobacteriales; Coriobacteriaceae; Collinsella;
|
| CAZyme ID |
MGYG000001588_00881
|
| CAZy Family |
GT83 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 794 |
|
79103.82 |
5.5524 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000001588 |
2411406 |
MAG |
Canada |
North America |
|
| Gene Location |
Start: 12353;
End: 14737
Strand: -
|
No EC number prediction in MGYG000001588_00881.
| Family |
Start |
End |
Evalue |
family coverage |
| GT83 |
60 |
534 |
3.9e-40 |
0.7814814814814814 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| pfam13231
|
PMT_2 |
3.48e-21 |
117 |
262 |
1 |
137 |
Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
| COG1807
|
ArnT |
9.98e-16 |
112 |
432 |
58 |
321 |
4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
|
O34575
|
1.05e-34 |
67 |
325 |
15 |
265 |
Putative mannosyltransferase YkcB OS=Bacillus subtilis (strain 168) OX=224308 GN=ykcB PE=3 SV=2 |
|
P37483
|
1.17e-34 |
62 |
325 |
8 |
263 |
Putative mannosyltransferase YycA OS=Bacillus subtilis (strain 168) OX=224308 GN=yycA PE=3 SV=2 |
This protein is predicted as OTHER
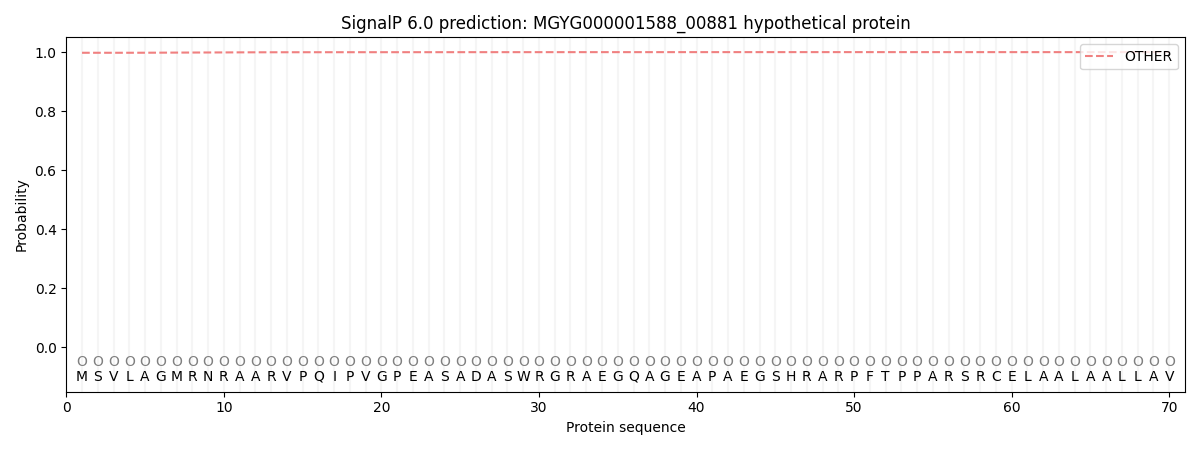
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.998044
|
0.001793
|
0.000112
|
0.000009
|
0.000006
|
0.000038
|
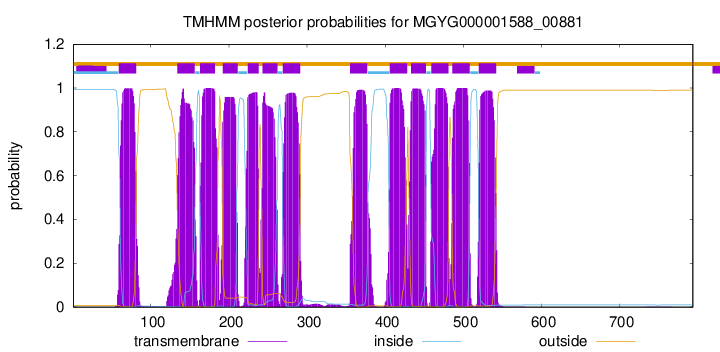
| start |
end |
| 59 |
81 |
| 134 |
156 |
| 163 |
182 |
| 192 |
211 |
| 224 |
238 |
| 243 |
262 |
| 269 |
291 |
| 355 |
377 |
| 406 |
428 |
| 433 |
452 |
| 459 |
481 |
| 486 |
508 |
| 520 |
542 |


