You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001591_01468
You are here: Home > Sequence: MGYG000001591_01468
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parabacteroides timonensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides timonensis | |||||||||||
| CAZyme ID | MGYG000001591_01468 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 276; End: 3446 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 22 | 612 | 9.4e-65 | 0.586436170212766 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 6.01e-28 | 28 | 568 | 13 | 515 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10340 | ebgA | 7.25e-18 | 25 | 469 | 39 | 471 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| PRK10150 | PRK10150 | 2.35e-16 | 29 | 468 | 14 | 443 | beta-D-glucuronidase; Provisional |
| pfam02837 | Glyco_hydro_2_N | 2.81e-10 | 28 | 182 | 2 | 157 | Glycosyl hydrolases family 2, sugar binding domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities and has a jelly-roll fold. The domain binds the sugar moiety during the sugar-hydrolysis reaction. |
| pfam00703 | Glyco_hydro_2 | 3.52e-09 | 201 | 303 | 6 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QQD16168.1 | 0.0 | 9 | 1056 | 8 | 1075 |
| QQT56131.1 | 0.0 | 5 | 1056 | 6 | 1074 |
| QTE22603.1 | 0.0 | 8 | 1056 | 6 | 1066 |
| QOD61956.1 | 0.0 | 5 | 1056 | 1 | 1066 |
| AOM80476.1 | 0.0 | 9 | 1055 | 6 | 1049 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6S6Z_A | 2.85e-22 | 29 | 477 | 40 | 471 | Structureof beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_B Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_C Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_D Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_E Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_F Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_G Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_H Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8] |
| 6SD0_A | 2.86e-22 | 29 | 477 | 41 | 472 | Structureof beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_B Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_C Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_D Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8] |
| 6ECA_A | 9.47e-18 | 24 | 526 | 37 | 535 | Lactobacillusrhamnosus Beta-glucuronidase [Lacticaseibacillus rhamnosus],6ECA_B Lactobacillus rhamnosus Beta-glucuronidase [Lacticaseibacillus rhamnosus] |
| 5T9A_A | 1.23e-14 | 18 | 446 | 21 | 435 | Crystalstructure of BuGH2Cwt [Bacteroides uniformis],5T9A_B Crystal structure of BuGH2Cwt [Bacteroides uniformis],5T9A_C Crystal structure of BuGH2Cwt [Bacteroides uniformis],5T9A_D Crystal structure of BuGH2Cwt [Bacteroides uniformis],5T9G_A Crystal structure of BuGH2Cwt in complex with Galactoisofagomine [Bacteroides uniformis],5T9G_B Crystal structure of BuGH2Cwt in complex with Galactoisofagomine [Bacteroides uniformis],5T9G_C Crystal structure of BuGH2Cwt in complex with Galactoisofagomine [Bacteroides uniformis],5T9G_D Crystal structure of BuGH2Cwt in complex with Galactoisofagomine [Bacteroides uniformis] |
| 6LEM_B | 1.88e-13 | 28 | 445 | 11 | 411 | ChainB, Beta-D-glucuronidase [Escherichia coli] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q56307 | 1.56e-21 | 29 | 477 | 41 | 472 | Beta-galactosidase OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=lacZ PE=1 SV=2 |
| A7LXS9 | 7.61e-15 | 19 | 457 | 36 | 442 | Beta-galactosidase BoGH2A OS=Bacteroides ovatus (strain ATCC 8483 / DSM 1896 / JCM 5824 / BCRC 10623 / CCUG 4943 / NCTC 11153) OX=411476 GN=BACOVA_02645 PE=1 SV=1 |
| Q6D736 | 9.95e-14 | 18 | 459 | 48 | 487 | Beta-galactosidase OS=Pectobacterium atrosepticum (strain SCRI 1043 / ATCC BAA-672) OX=218491 GN=lacZ PE=3 SV=1 |
| A8AKB8 | 5.06e-13 | 22 | 459 | 50 | 477 | Beta-galactosidase OS=Citrobacter koseri (strain ATCC BAA-895 / CDC 4225-83 / SGSC4696) OX=290338 GN=lacZ PE=3 SV=1 |
| P05804 | 1.04e-12 | 28 | 445 | 13 | 413 | Beta-glucuronidase OS=Escherichia coli (strain K12) OX=83333 GN=uidA PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
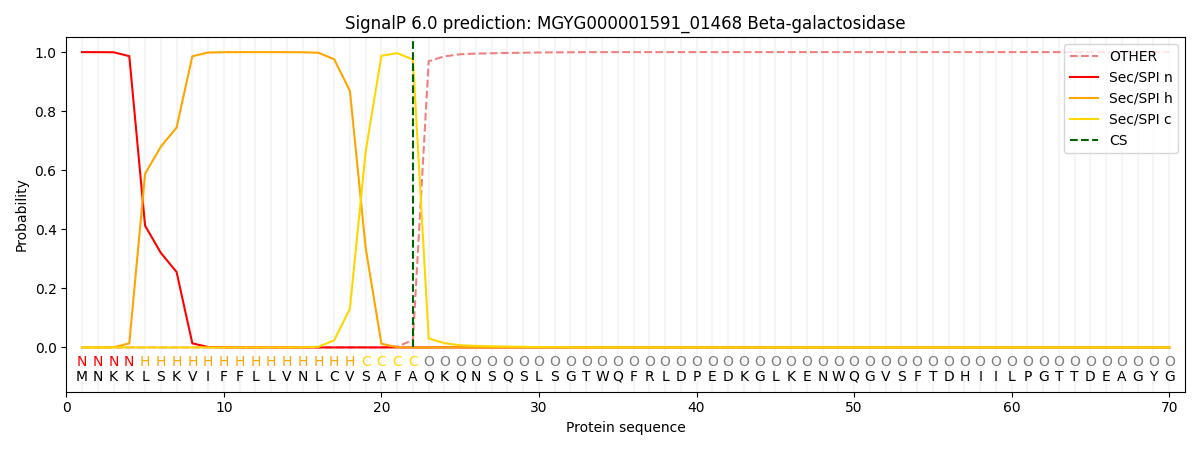
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000348 | 0.998656 | 0.000447 | 0.000187 | 0.000172 | 0.000163 |
