You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001592_00439
You are here: Home > Sequence: MGYG000001592_00439
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Intestinimonas sp900540545 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Oscillospiraceae; Intestinimonas; Intestinimonas sp900540545 | |||||||||||
| CAZyme ID | MGYG000001592_00439 | |||||||||||
| CAZy Family | GH36 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 206430; End: 209630 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH36 | 299 | 651 | 9.7e-32 | 0.5247093023255814 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd14791 | GH36 | 1.12e-33 | 299 | 560 | 9 | 260 | glycosyl hydrolase family 36 (GH36). GH36 enzymes occur in prokaryotes, eukaryotes, and archaea with a wide range of hydrolytic activities, including alpha-galactosidase, alpha-N-acetylgalactosaminidase, stachyose synthase, and raffinose synthase. All GH36 enzymes cleave a terminal carbohydrate moiety from a substrate that varies considerably in size, depending on the enzyme, and may be either a starch or a glycoprotein. GH36 members are retaining enzymes that cleave their substrates via an acid/base-catalyzed, double-displacement mechanism involving a covalent glycosyl-enzyme intermediate. Two aspartic acid residues have been identified as the catalytic nucleophile and the acid/base, respectively. |
| COG3345 | GalA | 1.60e-14 | 286 | 394 | 283 | 396 | Alpha-galactosidase [Carbohydrate transport and metabolism]. |
| pfam02065 | Melibiase | 6.20e-14 | 281 | 391 | 27 | 142 | Melibiase. Glycoside hydrolase families GH27, GH31 and GH36 form the glycoside hydrolase clan GH-D. Glycoside hydrolase family 36 can be split into 11 families, GH36A to GH36K. This family includes enzymes from GH36A-B and GH36D-K and from GH27. |
| cd06592 | GH31_NET37 | 5.12e-09 | 299 | 487 | 5 | 183 | glucosidase NET37. NET37 (also known as KIAA1161) is a human lamina-associated nuclear envelope transmembrane protein. A member of the glycosyl hydrolase family 31 (GH31) , it has been shown to be required for myogenic differentiation of C2C12 cells. Related proteins are found in eukaryotes and prokaryotes. Enzymes of the GH31 family possess a wide range of different hydrolytic activities including alpha-glucosidase (glucoamylase and sucrase-isomaltase), alpha-xylosidase, 6-alpha-glucosyltransferase, 3-alpha-isomaltosyltransferase and alpha-1,4-glucan lyase. All GH31 enzymes cleave a terminal carbohydrate moiety from a substrate that varies considerably in size, depending on the enzyme, and may be either a starch or a glycoprotein. |
| pfam17996 | CE2_N | 1.74e-05 | 962 | 1054 | 1 | 84 | Carbohydrate esterase 2 N-terminal. This is the N-terminal beta-sheet domain with jelly roll topology found in CE2 acetyl-esterase from the bacterium Clostridium thermocellum. This enzyme displays dual activities, it catalyses the deacetylation of plant polysaccharides and also potentiates the activity of its appended cellulase catalytic module through its noncatalytic cellulose binding function. This N-terminal jelly-roll domain appears to extend the substrate/cellulose binding cleft of the catalytic domain in C.thermocellum. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIA30399.1 | 0.0 | 1 | 1066 | 1 | 1076 |
| QQR05374.1 | 0.0 | 1 | 1066 | 1 | 1076 |
| ANU41754.1 | 0.0 | 1 | 1066 | 1 | 1076 |
| ABF39669.1 | 4.00e-89 | 26 | 680 | 31 | 681 |
| ARU29327.1 | 1.12e-80 | 70 | 683 | 73 | 689 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2XN0_A | 5.45e-11 | 298 | 393 | 339 | 436 | Structureof alpha-galactosidase from Lactobacillus acidophilus NCFM, PtCl4 derivative [Lactobacillus acidophilus NCFM],2XN0_B Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM, PtCl4 derivative [Lactobacillus acidophilus NCFM],2XN1_A Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM with TRIS [Lactobacillus acidophilus NCFM],2XN1_B Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM with TRIS [Lactobacillus acidophilus NCFM],2XN1_C Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM with TRIS [Lactobacillus acidophilus NCFM],2XN1_D Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM with TRIS [Lactobacillus acidophilus NCFM] |
| 2XN2_A | 9.42e-11 | 298 | 391 | 339 | 434 | Structureof alpha-galactosidase from Lactobacillus acidophilus NCFM with galactose [Lactobacillus acidophilus NCFM] |
| 2YFN_A | 1.28e-08 | 267 | 453 | 310 | 479 | galactosidasedomain of alpha-galactosidase-sucrose kinase, AgaSK [[Ruminococcus] gnavus E1],2YFO_A GALACTOSIDASE DOMAIN OF ALPHA-GALACTOSIDASE-SUCROSE KINASE, AGASK, in complex with galactose [[Ruminococcus] gnavus E1] |
| 6PHU_A | 2.94e-08 | 306 | 394 | 355 | 447 | SpAgawild type apo structure [Streptococcus pneumoniae TIGR4],6PHV_A Chain A, Alpha-galactosidase [Streptococcus pneumoniae TIGR4] |
| 6PHW_A | 2.94e-08 | 306 | 394 | 355 | 447 | ChainA, Alpha-galactosidase [Streptococcus pneumoniae TIGR4],6PHX_A SpAga D472N structure in complex with raffinose [Streptococcus pneumoniae TIGR4],6PHY_A Chain A, Alpha-galactosidase [Streptococcus pneumoniae TIGR4],6PI0_A AgaD472N-Linear Blood group B type 2 trisaccharide complex structure [Streptococcus pneumoniae TIGR4] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0DTR5 | 0.0 | 1 | 1066 | 1 | 1076 | A type blood alpha-D-galactosamine galactosaminidase OS=Flavonifractor plautii OX=292800 PE=1 SV=1 |
| P0DTR4 | 8.75e-39 | 929 | 1066 | 638 | 771 | A type blood N-acetyl-alpha-D-galactosamine deacetylase OS=Flavonifractor plautii OX=292800 PE=1 SV=1 |
| G1UB44 | 2.98e-10 | 298 | 393 | 339 | 436 | Alpha-galactosidase Mel36A OS=Lactobacillus acidophilus (strain ATCC 700396 / NCK56 / N2 / NCFM) OX=272621 GN=melA PE=1 SV=1 |
| P27756 | 1.03e-08 | 285 | 394 | 313 | 427 | Alpha-galactosidase OS=Streptococcus mutans serotype c (strain ATCC 700610 / UA159) OX=210007 GN=aga PE=3 SV=3 |
| Q92457 | 7.12e-08 | 244 | 519 | 302 | 568 | Alpha-galactosidase 2 OS=Hypocrea jecorina OX=51453 GN=agl2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
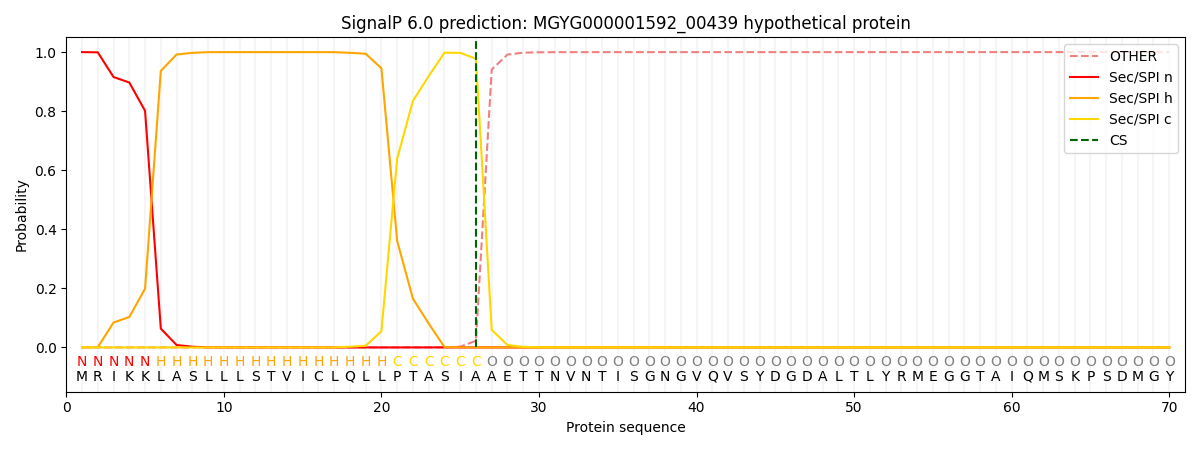
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000215 | 0.999004 | 0.000277 | 0.000176 | 0.000161 | 0.000149 |
