You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001594_01482
You are here: Home > Sequence: MGYG000001594_01482
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
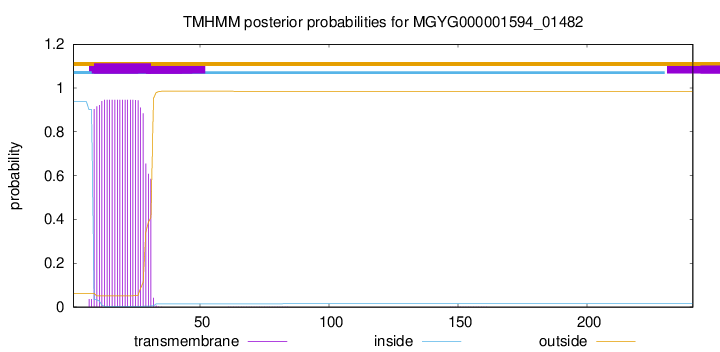
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Blautia_A; | |||||||||||
| CAZyme ID | MGYG000001594_01482 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 4504; End: 5229 Strand: - | |||||||||||
Full Sequence Download help
| MIFSRKNKLF WVNLTLTGAL IFGAPVFVQG AADHSGEGQI HKPSYNITTG PGTEGFIDDS | 60 |
| TDNTYGYYTI MGTTTVTLEE MTDLYETQGA EYPQSLKEGG AETIEDFCSI ILEEAEEEGV | 120 |
| RAEVVYVQAM LETGWLQFGG DAVESQFNFS GLGTTGGGVQ GNSYPDVRTG IRAQVQHLKA | 180 |
| YASTEELSGN CVDSRFDMVK RGSAPYVEWL GIQENPNGGG WAAGAGYGEK LRKLLTDLKG | 240 |
| E | 241 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH73 | 109 | 236 | 1.5e-18 | 0.9453125 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01832 | Glucosaminidase | 2.97e-09 | 108 | 174 | 1 | 77 | Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase. This family includes Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase EC:3.2.1.96. As well as the flageller protein J that has been shown to hydrolyze peptidoglycan. |
| PRK12711 | flgJ | 0.003 | 100 | 186 | 211 | 304 | flagellar assembly peptidoglycan hydrolase FlgJ. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBL24117.1 | 7.95e-109 | 1 | 241 | 1 | 274 |
| CBL19417.1 | 1.59e-103 | 14 | 241 | 2 | 229 |
| QCU04040.1 | 8.68e-101 | 14 | 241 | 2 | 229 |
| QHB24702.1 | 8.28e-69 | 66 | 241 | 84 | 260 |
| QEI32209.1 | 8.28e-69 | 66 | 241 | 84 | 260 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O51481 | 2.02e-25 | 112 | 238 | 76 | 195 | Uncharacterized protein BB_0531 OS=Borreliella burgdorferi (strain ATCC 35210 / DSM 4680 / CIP 102532 / B31) OX=224326 GN=BB_0531 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.107949 | 0.888893 | 0.001906 | 0.000507 | 0.000354 | 0.000357 |