You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001596_01626
You are here: Home > Sequence: MGYG000001596_01626
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Blautia sp900543715 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Blautia; Blautia sp900543715 | |||||||||||
| CAZyme ID | MGYG000001596_01626 | |||||||||||
| CAZy Family | CE1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 11133; End: 12206 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE1 | 113 | 349 | 1.2e-28 | 0.9162995594713657 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG2382 | Fes | 1.96e-13 | 94 | 264 | 57 | 210 | Enterochelin esterase or related enzyme [Inorganic ion transport and metabolism]. |
| pfam00756 | Esterase | 0.001 | 113 | 241 | 9 | 120 | Putative esterase. This family contains Esterase D. However it is not clear if all members of the family have the same function. This family is related to the pfam00135 family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADA67819.1 | 6.37e-103 | 69 | 310 | 36 | 277 |
| AEY68182.1 | 5.91e-67 | 76 | 357 | 407 | 686 |
| ACL77751.1 | 7.13e-63 | 76 | 357 | 412 | 691 |
| AGA59284.1 | 7.68e-63 | 76 | 352 | 316 | 591 |
| QNU67689.1 | 1.56e-62 | 76 | 352 | 322 | 596 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1GKL_A | 2.27e-54 | 112 | 352 | 53 | 283 | ChainA, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],1GKL_B Chain B, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],1WB4_A Chain A, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus],1WB4_B Chain B, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus],1WB5_A Chain A, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus],1WB5_B Chain B, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus],1WB6_A Chain A, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],1WB6_B Chain B, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus] |
| 6FJ4_A | 1.11e-50 | 112 | 352 | 39 | 269 | ChainA, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus] |
| 1GKK_A | 1.63e-50 | 112 | 352 | 53 | 283 | ChainA, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus],1GKK_B Chain B, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus],3ZI7_A Chain A, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus YS],3ZI7_B Chain B, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus YS],4BAG_A Chain A, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],4BAG_B Chain B, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],4H35_A Chain A, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus],4H35_B Chain B, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus],5FXM_A Chain A, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus],6Y8G_AAA Chain AAA, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus],6Y8G_BBB Chain BBB, Endo-1,4-beta-xylanase Y [Acetivibrio thermocellus] |
| 1JJF_A | 1.50e-18 | 114 | 263 | 48 | 177 | ChainA, Endo-1,4-beta-xylanase Z [Acetivibrio thermocellus] |
| 1JT2_A | 3.86e-18 | 114 | 263 | 48 | 177 | ChainA, PROTEIN (ENDO-1,4-BETA-XYLANASE Z) [Acetivibrio thermocellus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P51584 | 1.33e-48 | 112 | 352 | 841 | 1071 | Endo-1,4-beta-xylanase Y OS=Acetivibrio thermocellus OX=1515 GN=xynY PE=1 SV=1 |
| P10478 | 7.54e-17 | 114 | 263 | 67 | 196 | Endo-1,4-beta-xylanase Z OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=xynZ PE=1 SV=3 |
| D5EY13 | 1.41e-10 | 114 | 279 | 516 | 664 | Endo-1,4-beta-xylanase/feruloyl esterase OS=Prevotella ruminicola (strain ATCC 19189 / JCM 8958 / 23) OX=264731 GN=xyn10D-fae1A PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
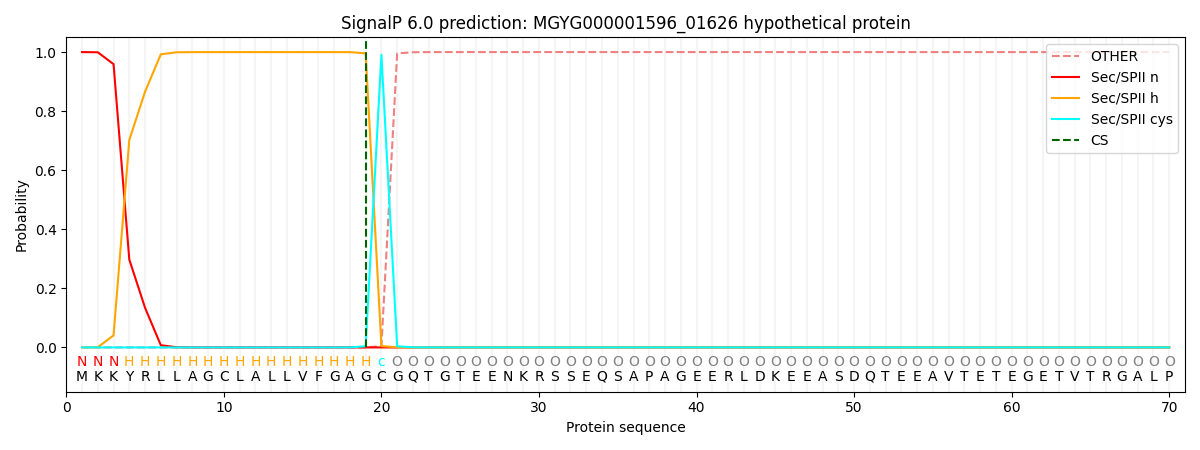
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000051 | 0.000000 | 0.000000 | 0.000000 |
