You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001610_01261
You are here: Home > Sequence: MGYG000001610_01261
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
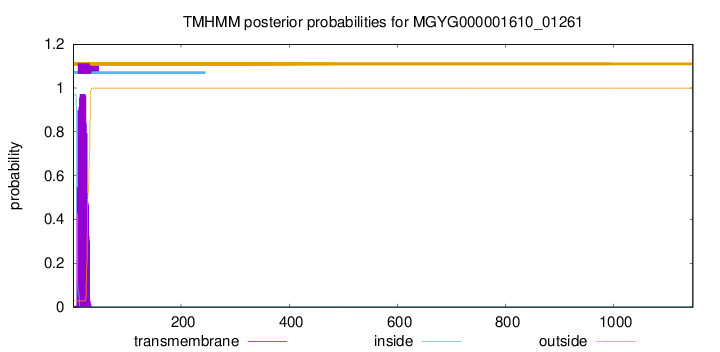
TMHMM annotations
Basic Information help
| Species | Sporanaerobacter acetigenes | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Tissierellales; Sporanaerobacteraceae; Sporanaerobacter; Sporanaerobacter acetigenes | |||||||||||
| CAZyme ID | MGYG000001610_01261 | |||||||||||
| CAZy Family | CBM50 | |||||||||||
| CAZyme Description | Trifunctional nucleotide phosphoesterase protein YfkN | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 88096; End: 91539 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK09419 | PRK09419 | 0.0 | 1 | 1081 | 2 | 1146 | multifunctional 2',3'-cyclic-nucleotide 2'-phosphodiesterase/3'-nucleotidase/5'-nucleotidase. |
| COG0737 | UshA | 4.54e-124 | 606 | 1072 | 24 | 503 | 2',3'-cyclic-nucleotide 2'-phosphodiesterase/5'- or 3'-nucleotidase, 5'-nucleotidase family [Nucleotide transport and metabolism, Defense mechanisms]. |
| PRK09558 | ushA | 3.34e-117 | 609 | 1071 | 35 | 538 | bifunctional UDP-sugar hydrolase/5'-nucleotidase periplasmic precursor; Reviewed |
| COG0737 | UshA | 3.05e-101 | 14 | 502 | 1 | 501 | 2',3'-cyclic-nucleotide 2'-phosphodiesterase/5'- or 3'-nucleotidase, 5'-nucleotidase family [Nucleotide transport and metabolism, Defense mechanisms]. |
| PRK09420 | cpdB | 3.08e-90 | 22 | 544 | 10 | 597 | bifunctional 2',3'-cyclic-nucleotide 2'-phosphodiesterase/3'-nucleotidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUI23903.1 | 3.49e-294 | 40 | 1075 | 85 | 1119 |
| VDN48620.1 | 3.52e-278 | 34 | 1069 | 83 | 1118 |
| QQY79959.1 | 6.64e-254 | 609 | 1146 | 39 | 576 |
| QGU94268.1 | 1.70e-239 | 2 | 1147 | 4 | 1147 |
| QZY54554.1 | 1.98e-211 | 607 | 1146 | 32 | 574 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2Z1A_A | 8.30e-74 | 609 | 1094 | 30 | 548 | Crystalstructure of 5'-nucleotidase precursor from Thermus thermophilus HB8 [Thermus thermophilus HB8] |
| 4H2F_A | 3.60e-68 | 606 | 1078 | 23 | 543 | Humanecto-5'-nucleotidase (CD73): crystal form I (open) in complex with adenosine [Homo sapiens],4H2G_A Human ecto-5'-nucleotidase (CD73): crystal form II (open) in complex with adenosine [Homo sapiens] |
| 4H1Y_P | 4.91e-68 | 606 | 1078 | 23 | 543 | Humanecto-5'-nucleotidase (CD73): crystal form II (open) in complex with PSB11552 [Homo sapiens],6TVE_P Unliganded human CD73 (5'-nucleotidase) in the open state [Homo sapiens],6TVG_A Human CD73 (ecto 5'-nucleotidase) in complex with AMPCP in the open state [Homo sapiens],7BBJ_A Chain A, 5'-nucleotidase [Homo sapiens],7BBJ_B Chain B, 5'-nucleotidase [Homo sapiens],7P9N_A Chain A, 5'-nucleotidase [Homo sapiens],7P9R_A Chain A, 5'-nucleotidase [Homo sapiens],7P9T_A Chain A, 5'-nucleotidase [Homo sapiens],7PA4_A Chain A, 5'-nucleotidase [Homo sapiens],7PB5_A Chain A, 5'-nucleotidase [Homo sapiens],7PBA_A Chain A, 5'-nucleotidase [Homo sapiens],7PBB_A Chain A, 5'-nucleotidase [Homo sapiens],7PBY_A Chain A, 5'-nucleotidase [Homo sapiens],7PCP_A Chain A, 5'-nucleotidase [Homo sapiens],7PD9_A Chain A, 5'-nucleotidase [Homo sapiens] |
| 4H2B_A | 5.02e-68 | 606 | 1078 | 24 | 544 | Humanecto-5'-nucleotidase (CD73): crystal form II (open) in complex with Baicalin [Homo sapiens] |
| 3IVD_A | 7.03e-68 | 607 | 1076 | 5 | 494 | Putative5'-Nucleotidase (c4898) from Escherichia Coli in complex with Uridine [Escherichia coli O6],3IVD_B Putative 5'-Nucleotidase (c4898) from Escherichia Coli in complex with Uridine [Escherichia coli O6],3IVE_A Putative 5'-Nucleotidase (c4898) from Escherichia Coli in complex with Cytidine [Escherichia coli O6] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O34313 | 4.26e-174 | 22 | 1081 | 26 | 1177 | Trifunctional nucleotide phosphoesterase protein YfkN OS=Bacillus subtilis (strain 168) OX=224308 GN=yfkN PE=1 SV=1 |
| A9BJC1 | 6.88e-82 | 606 | 1071 | 21 | 488 | Mannosylglucosyl-3-phosphoglycerate phosphatase OS=Petrotoga mobilis (strain DSM 10674 / SJ95) OX=403833 GN=mggB PE=1 SV=1 |
| Q05927 | 1.91e-69 | 606 | 1078 | 26 | 546 | 5'-nucleotidase OS=Bos taurus OX=9913 GN=NT5E PE=1 SV=2 |
| F8S0Z7 | 9.48e-67 | 608 | 1078 | 43 | 561 | Snake venom 5'-nucleotidase OS=Crotalus adamanteus OX=8729 PE=1 SV=2 |
| P21588 | 9.91e-67 | 608 | 1078 | 30 | 548 | 5'-nucleotidase OS=Rattus norvegicus OX=10116 GN=Nt5e PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
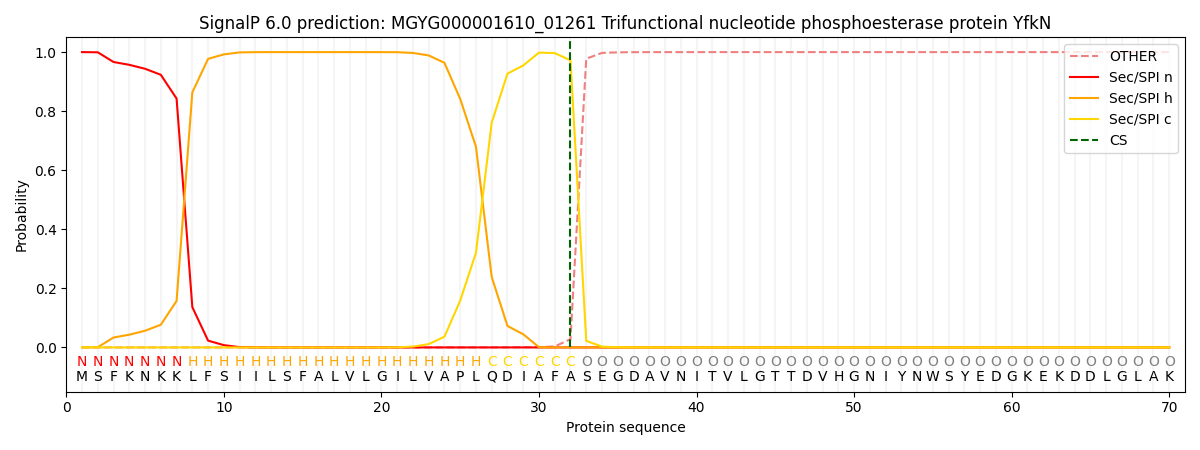
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000308 | 0.998994 | 0.000176 | 0.000184 | 0.000164 | 0.000151 |