You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001621_00384
You are here: Home > Sequence: MGYG000001621_00384
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pauljensenia sp900554605 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Actinomycetaceae; Pauljensenia; Pauljensenia sp900554605 | |||||||||||
| CAZyme ID | MGYG000001621_00384 | |||||||||||
| CAZy Family | GH66 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 464; End: 3121 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH66 | 39 | 400 | 6e-91 | 0.6618705035971223 |
| CBM35 | 396 | 526 | 4.3e-25 | 0.9915966386554622 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13199 | Glyco_hydro_66 | 1.04e-137 | 43 | 717 | 2 | 557 | Glycosyl hydrolase family 66. This family is a set of glycosyl hydrolase enzymes including cycloisomaltooligosaccharide glucanotransferase (EC:2.4.1.-) and dextranase (EC:3.2.1.11) activities. |
| cd14745 | GH66 | 9.78e-124 | 126 | 588 | 1 | 331 | Glycoside Hydrolase Family 66. Glycoside Hydrolase Family 66 contains proteins characterized as cycloisomaltooligosaccharide glucanotransferase (CITase) and dextranases from a variety of bacteria. CITase cyclizes part of a (1-6)-alpha-D-glucan (dextrans) chain by formation of a (1-6)-alpha-D-glucosidic bond. Dextranases catalyze the endohydrolysis of (1-6)-alpha-D-glucosidic linkages in dextran. Some members contain Carbohydrate Binding Module 35 (CBM35) domains, either C-terminal or inserted in the domain or both. |
| cd04083 | CBM35_Lmo2446-like | 2.35e-26 | 395 | 526 | 2 | 125 | Carbohydrate Binding Module 35 (CBM35) domains similar to Lmo2446. This family includes carbohydrate binding module 35 (CBM35) domains that are appended to several carbohydrate binding enzymes. Some CBM35 domains belonging to this family are appended to glycoside hydrolase (GH) family domains, including glycoside hydrolase family 31 (GH31), for example the CBM35 domain of Lmo2446, an uncharacterized protein from Listeria monocytogenes EGD-e. These CBM35s are non-catalytic carbohydrate binding domains that facilitate the strong binding of the GH catalytic modules with their dedicated, insoluble substrates. GH31 has a wide range of hydrolytic activities such as alpha-glucosidase, alpha-xylosidase, 6-alpha-glucosyltransferase, or alpha-1,4-glucan lyase, cleaving a terminal carbohydrate moiety from a substrate that may be a starch or a glycoprotein. Most characterized GH31 enzymes are alpha-glucosidases. |
| pfam00652 | Ricin_B_lectin | 9.39e-15 | 751 | 881 | 3 | 126 | Ricin-type beta-trefoil lectin domain. |
| NF035929 | lectin_1 | 1.42e-14 | 758 | 882 | 721 | 835 | lectin. Lectins are important adhesin proteins, which bind carbohydrate structures on host cell surface. The carbohydrate specificity of diverse lectins to a large extent dictates bacteria tissue tropism by mediating specific attachment to unique host sites expressing the corresponding carbohydrate receptor. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGS11685.1 | 1.62e-190 | 2 | 801 | 4 | 767 |
| QIS74997.1 | 7.60e-159 | 37 | 782 | 38 | 649 |
| QCT35775.1 | 1.40e-148 | 37 | 851 | 40 | 901 |
| QNK58819.1 | 1.75e-134 | 37 | 720 | 40 | 736 |
| BAA09604.1 | 2.84e-133 | 37 | 720 | 44 | 740 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3WNK_A | 2.09e-137 | 37 | 717 | 27 | 720 | ChainA, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans] |
| 3WNP_A | 2.32e-136 | 37 | 717 | 8 | 701 | ChainA, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans],3WNP_B Chain B, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans] |
| 3WNL_A | 2.32e-136 | 37 | 717 | 8 | 701 | ChainA, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans],3WNM_A Chain A, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans],3WNN_A Chain A, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans],3WNN_B Chain B, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans],3WNO_A Chain A, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans],3WNO_B Chain B, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans] |
| 5X7G_A | 5.44e-133 | 37 | 717 | 26 | 719 | CrystalStructure of Paenibacillus sp. 598K cycloisomaltooligosaccharide glucanotransferase [Paenibacillus sp. 598K],5X7H_A Crystal Structure of Paenibacillus sp. 598K cycloisomaltooligosaccharide glucanotransferase complexed with cycloisomaltoheptaose [Paenibacillus sp. 598K] |
| 5AXG_A | 1.73e-53 | 37 | 720 | 44 | 610 | Crystalstructure of thermophilic dextranase from Thermoanaerobacter pseudethanolicus [Thermoanaerobacter pseudethanolicus ATCC 33223],5AXG_B Crystal structure of thermophilic dextranase from Thermoanaerobacter pseudethanolicus [Thermoanaerobacter pseudethanolicus ATCC 33223] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P94286 | 5.67e-134 | 37 | 720 | 44 | 740 | Cycloisomaltooligosaccharide glucanotransferase OS=Niallia circulans OX=1397 PE=1 SV=1 |
| P70873 | 7.63e-131 | 37 | 724 | 36 | 736 | Cycloisomaltooligosaccharide glucanotransferase OS=Niallia circulans OX=1397 GN=cit PE=3 SV=1 |
| Q59979 | 7.84e-13 | 98 | 391 | 42 | 362 | Dextranase OS=Streptococcus salivarius OX=1304 GN=dex PE=3 SV=1 |
| P39653 | 1.70e-10 | 102 | 717 | 242 | 799 | Dextranase OS=Streptococcus downei OX=1317 GN=dex PE=1 SV=1 |
| Q54443 | 2.50e-10 | 102 | 410 | 184 | 518 | Dextranase OS=Streptococcus mutans serotype c (strain ATCC 700610 / UA159) OX=210007 GN=dexA PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
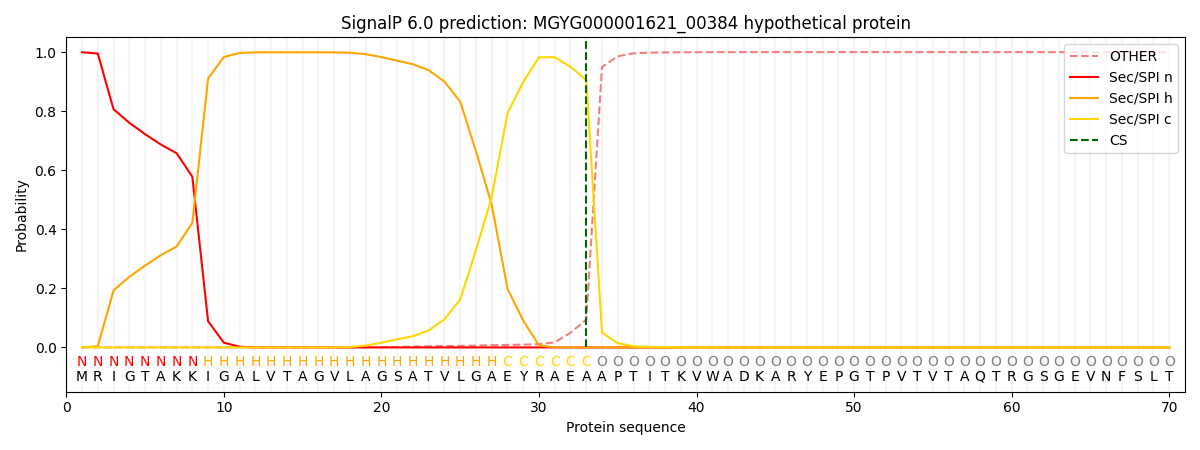
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001806 | 0.996623 | 0.000351 | 0.000632 | 0.000304 | 0.000229 |
