You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001622_01031
You are here: Home > Sequence: MGYG000001622_01031
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Enterocloster sp900549235 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Enterocloster; Enterocloster sp900549235 | |||||||||||
| CAZyme ID | MGYG000001622_01031 | |||||||||||
| CAZy Family | CBM50 | |||||||||||
| CAZyme Description | Trifunctional nucleotide phosphoesterase protein YfkN | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 178939; End: 180759 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK09419 | PRK09419 | 2.40e-167 | 49 | 600 | 42 | 644 | multifunctional 2',3'-cyclic-nucleotide 2'-phosphodiesterase/3'-nucleotidase/5'-nucleotidase. |
| COG0737 | UshA | 6.93e-90 | 47 | 505 | 25 | 499 | 2',3'-cyclic-nucleotide 2'-phosphodiesterase/5'- or 3'-nucleotidase, 5'-nucleotidase family [Nucleotide transport and metabolism, Defense mechanisms]. |
| cd07410 | MPP_CpdB_N | 3.33e-82 | 49 | 308 | 1 | 276 | Escherichia coli CpdB and related proteins, N-terminal metallophosphatase domain. CpdB is a bacterial periplasmic protein with an N-terminal metallophosphatase domain and a C-terminal 3'-nucleotidase domain. This alignment model represents the N-terminal metallophosphatase domain, which has 2',3'-cyclic phosphodiesterase activity, hydrolyzing the 2',3'-cyclic phosphates of adenosine, guanosine, cytosine and uridine to yield nucleoside and phosphate. CpdB also hydrolyzes the chromogenic substrates p-nitrophenyl phosphate (PNPP), bis(PNPP) and p-nitrophenyl phosphorylcholine (NPPC). CpdB is thought to play a scavenging role during RNA hydrolysis by converting the non-transportable nucleotides produced by RNaseI to nucleosides which can easily enter a cell for use as a carbon source. This family also includes YfkN, a Bacillus subtilis nucleotide phosphoesterase with two copies of each of the metallophosphatase and 3'-nucleotidase domains. The N-terminal metallophosphatase domain belongs to a large superfamily of distantly related metallophosphatases (MPPs) that includes: Mre11/SbcD-like exonucleases, Dbr1-like RNA lariat debranching enzymes, YfcE-like phosphodiesterases, purple acid phosphatases (PAPs), YbbF-like UDP-2,3-diacylglucosamine hydrolases, and acid sphingomyelinases (ASMases). MPPs are functionally diverse, but all share a conserved domain with an active site consisting of two metal ions (usually manganese, iron, or zinc) coordinated with octahedral geometry by a cage of histidine, aspartate, and asparagine residues. The conserved domain is a double beta-sheet sandwich with a di-metal active site made up of residues located at the C-terminal side of the sheets. This domain is thought to allow for productive metal coordination. |
| PRK09420 | cpdB | 4.22e-70 | 48 | 580 | 25 | 624 | bifunctional 2',3'-cyclic-nucleotide 2'-phosphodiesterase/3'-nucleotidase. |
| PRK09418 | PRK09418 | 3.06e-66 | 47 | 559 | 38 | 605 | bifunctional 2',3'-cyclic-nucleotide 2'-phosphodiesterase/3'-nucleotidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CDI50173.1 | 2.85e-224 | 43 | 605 | 32 | 606 |
| AAO36497.1 | 2.14e-221 | 43 | 605 | 32 | 606 |
| QBD85406.1 | 8.62e-221 | 43 | 605 | 32 | 606 |
| AVP54978.1 | 1.22e-220 | 43 | 605 | 32 | 606 |
| SNV84369.1 | 3.83e-214 | 43 | 605 | 32 | 606 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3GVE_A | 5.71e-38 | 41 | 304 | 4 | 305 | Crystalstructure of calcineurin-like phosphoesterase YfkN from Bacillus subtilis [Bacillus subtilis subsp. subtilis str. 168],3GVE_B Crystal structure of calcineurin-like phosphoesterase YfkN from Bacillus subtilis [Bacillus subtilis subsp. subtilis str. 168] |
| 5EQV_A | 6.46e-33 | 43 | 309 | 4 | 304 | 1.45Angstrom Crystal Structure of Bifunctional 2',3'-cyclic Nucleotide 2'-phosphodiesterase/3'-Nucleotidase Periplasmic Precursor Protein from Yersinia pestis with Phosphate bound to the Active site [Yersinia pestis CO92] |
| 3QFK_A | 3.49e-31 | 48 | 530 | 19 | 501 | ChainA, Uncharacterized protein [Staphylococcus aureus subsp. aureus NCTC 8325] |
| 4Q7F_A | 3.49e-31 | 48 | 530 | 19 | 501 | ChainA, 5' nucleotidase family protein [Staphylococcus aureus subsp. aureus COL] |
| 3JYF_A | 9.35e-29 | 44 | 309 | 4 | 303 | ChainA, 2',3'-cyclic nucleotide 2'-phosphodiesterase/3'-nucleotidase bifunctional periplasmic protein [Klebsiella pneumoniae subsp. pneumoniae MGH 78578],3JYF_B Chain B, 2',3'-cyclic nucleotide 2'-phosphodiesterase/3'-nucleotidase bifunctional periplasmic protein [Klebsiella pneumoniae subsp. pneumoniae MGH 78578] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O34313 | 4.13e-58 | 41 | 561 | 37 | 609 | Trifunctional nucleotide phosphoesterase protein YfkN OS=Bacillus subtilis (strain 168) OX=224308 GN=yfkN PE=1 SV=1 |
| P08331 | 4.80e-51 | 44 | 580 | 19 | 622 | 2',3'-cyclic-nucleotide 2'-phosphodiesterase/3'-nucleotidase OS=Escherichia coli (strain K12) OX=83333 GN=cpdB PE=1 SV=2 |
| P53052 | 8.91e-50 | 44 | 580 | 24 | 627 | 2',3'-cyclic-nucleotide 2'-phosphodiesterase/3'-nucleotidase OS=Yersinia enterocolitica OX=630 GN=cpdB PE=3 SV=1 |
| P26265 | 6.25e-47 | 44 | 561 | 19 | 595 | 2',3'-cyclic-nucleotide 2'-phosphodiesterase/3'-nucleotidase OS=Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) OX=99287 GN=cpdB PE=3 SV=2 |
| P44764 | 5.57e-45 | 47 | 580 | 32 | 632 | 2',3'-cyclic-nucleotide 2'-phosphodiesterase/3'-nucleotidase OS=Haemophilus influenzae (strain ATCC 51907 / DSM 11121 / KW20 / Rd) OX=71421 GN=cpdB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
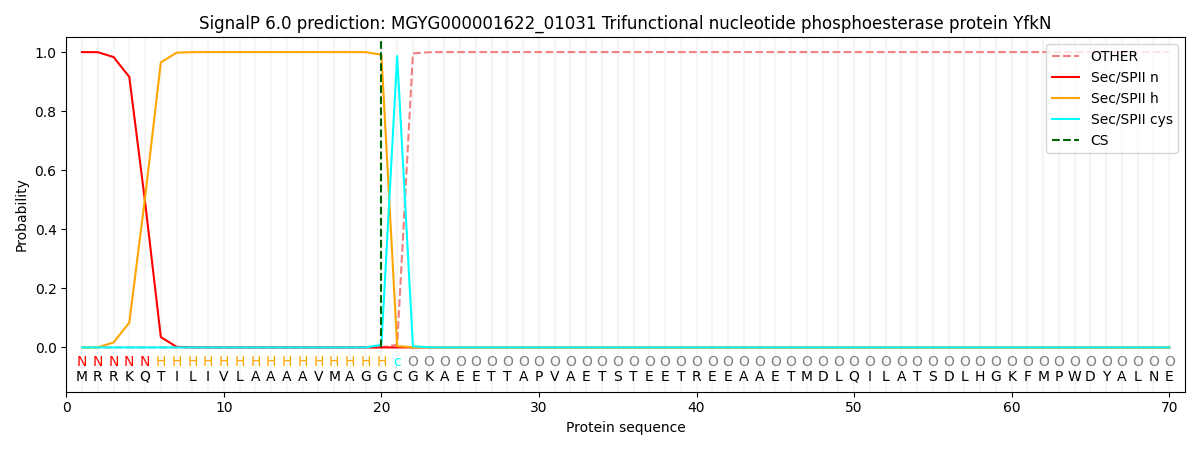
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000001 | 1.000049 | 0.000000 | 0.000000 | 0.000000 |
