You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001633_00303
You are here: Home > Sequence: MGYG000001633_00303
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | RC9 sp000435075 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; UBA932; RC9; RC9 sp000435075 | |||||||||||
| CAZyme ID | MGYG000001633_00303 | |||||||||||
| CAZy Family | GH63 | |||||||||||
| CAZyme Description | Glucosidase YgjK | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 156431; End: 158440 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH63 | 1 | 665 | 6.5e-150 | 0.987719298245614 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK10137 | PRK10137 | 6.56e-158 | 1 | 660 | 1 | 774 | alpha-glucosidase; Provisional |
| COG3408 | GDB1 | 5.77e-21 | 309 | 667 | 268 | 604 | Glycogen debranching enzyme (alpha-1,6-glucosidase) [Carbohydrate transport and metabolism]. |
| pfam01204 | Trehalase | 7.27e-20 | 381 | 666 | 178 | 505 | Trehalase. Trehalase (EC:3.2.1.28) is known to recycle trehalose to glucose. Trehalose is a physiological hallmark of heat-shock response in yeast and protects of proteins and membranes against a variety of stresses. This family is found in conjunction with pfam07492 in fungi. |
| COG1626 | TreA | 8.79e-14 | 473 | 666 | 354 | 549 | Neutral trehalase [Carbohydrate transport and metabolism]. |
| PLN02567 | PLN02567 | 2.12e-11 | 331 | 666 | 156 | 541 | alpha,alpha-trehalase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUU00825.1 | 0.0 | 7 | 667 | 8 | 667 |
| QQA30345.1 | 0.0 | 7 | 667 | 8 | 667 |
| QUT61859.1 | 0.0 | 7 | 667 | 8 | 667 |
| BBK86094.1 | 0.0 | 7 | 667 | 8 | 667 |
| ABR38022.1 | 5.55e-307 | 5 | 667 | 3 | 687 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3W7S_A | 1.25e-90 | 36 | 660 | 3 | 747 | Escherichiacoli K12 YgjK complexed with glucose [Escherichia coli K-12],3W7S_B Escherichia coli K12 YgjK complexed with glucose [Escherichia coli K-12],3W7T_A Escherichia coli K12 YgjK complexed with mannose [Escherichia coli K-12],3W7T_B Escherichia coli K12 YgjK complexed with mannose [Escherichia coli K-12],3W7U_A Escherichia coli K12 YgjK complexed with galactose [Escherichia coli K-12],3W7U_B Escherichia coli K12 YgjK complexed with galactose [Escherichia coli K-12] |
| 3W7X_A | 6.62e-90 | 36 | 660 | 3 | 747 | Crystalstructure of E. coli YgjK D324N complexed with melibiose [Escherichia coli K-12],3W7X_B Crystal structure of E. coli YgjK D324N complexed with melibiose [Escherichia coli K-12],5CA3_A Crystal structure of the glycosynthase mutant D324N of Escherichia coli GH63 glycosidase in complex with glucose and lactose [Escherichia coli K-12],5CA3_B Crystal structure of the glycosynthase mutant D324N of Escherichia coli GH63 glycosidase in complex with glucose and lactose [Escherichia coli K-12] |
| 3W7W_A | 9.24e-90 | 36 | 660 | 3 | 747 | Crystalstructure of E. coli YgjK E727A complexed with 2-O-alpha-D-glucopyranosyl-alpha-D-galactopyranose [Escherichia coli K-12],3W7W_B Crystal structure of E. coli YgjK E727A complexed with 2-O-alpha-D-glucopyranosyl-alpha-D-galactopyranose [Escherichia coli K-12],5GW7_A Crystal structure of the glycosynthase mutant E727A of Escherichia coli GH63 glycosidase in complex with glucose and lactose [Escherichia coli K-12],5GW7_B Crystal structure of the glycosynthase mutant E727A of Escherichia coli GH63 glycosidase in complex with glucose and lactose [Escherichia coli K-12] |
| 3D3I_A | 1.44e-85 | 36 | 660 | 4 | 748 | Crystalstructural of Escherichia coli K12 YgjK, a glucosidase belonging to glycoside hydrolase family 63 [Escherichia coli K-12],3D3I_B Crystal structural of Escherichia coli K12 YgjK, a glucosidase belonging to glycoside hydrolase family 63 [Escherichia coli K-12] |
| 7PQQ_B | 3.20e-43 | 439 | 660 | 41 | 296 | ChainB, Anti-RON nanobody,Megabody 91,Glucosidase YgjK [Lama glama] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P42592 | 1.11e-89 | 36 | 660 | 26 | 770 | Glucosidase YgjK OS=Escherichia coli (strain K12) OX=83333 GN=ygjK PE=1 SV=1 |
| D8QTR2 | 4.32e-26 | 329 | 669 | 98 | 488 | Mannosylglycerate hydrolase MGH1 OS=Selaginella moellendorffii OX=88036 GN=MGH PE=1 SV=1 |
| D8T3S4 | 4.32e-26 | 329 | 669 | 98 | 488 | Mannosylglycerate hydrolase MGH2 OS=Selaginella moellendorffii OX=88036 GN=SELMODRAFT_447962 PE=3 SV=1 |
| K5BDL0 | 3.38e-14 | 328 | 666 | 39 | 445 | Glucosylglycerate hydrolase OS=Mycolicibacterium hassiacum (strain DSM 44199 / CIP 105218 / JCM 12690 / 3849) OX=1122247 GN=ggh PE=1 SV=1 |
| P94250 | 7.67e-11 | 333 | 613 | 63 | 319 | Uncharacterized protein BB_0381 OS=Borreliella burgdorferi (strain ATCC 35210 / DSM 4680 / CIP 102532 / B31) OX=224326 GN=BB_0381 PE=4 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
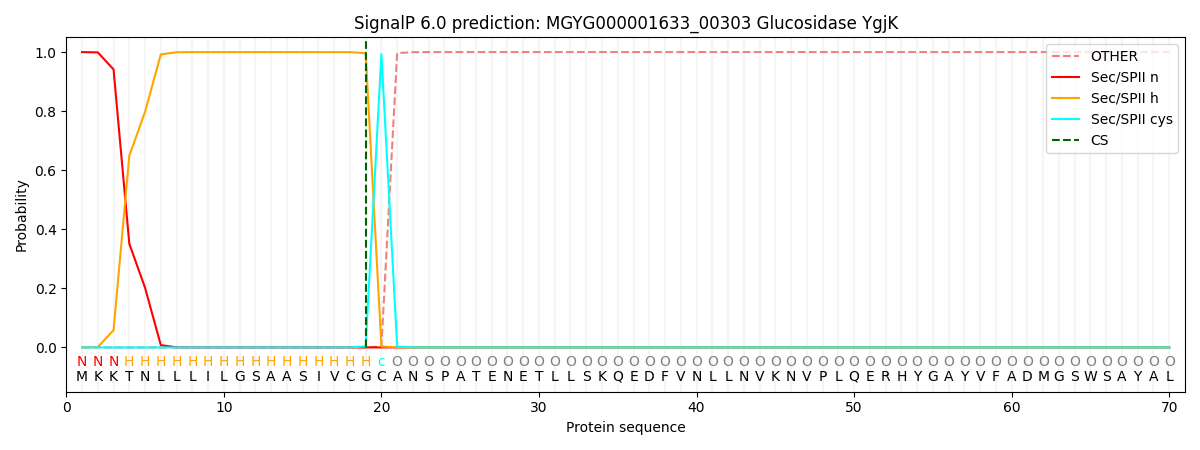
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000042 | 0.000000 | 0.000000 | 0.000000 |
