You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001639_00068
You are here: Home > Sequence: MGYG000001639_00068
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Haloferax massiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Archaea; Halobacteriota; Halobacteria; Halobacteriales; Haloferacaceae; Haloferax; Haloferax massiliensis | |||||||||||
| CAZyme ID | MGYG000001639_00068 | |||||||||||
| CAZy Family | AA1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 69657; End: 70811 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA1 | 111 | 347 | 2.2e-34 | 0.6564245810055865 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd04202 | CuRO_D2_2dMcoN_like | 1.18e-47 | 213 | 365 | 1 | 138 | The second cupredoxin domain of bacterial two domain multicopper oxidase McoN and similar proteins. This family includes bacterial two domain multicopper oxidases (2dMCOs) represented by the McoN from Nitrosomonas europaea. McoN is a trimeric type C blue copper oxidase. Each subunit houses a type 1 copper site in domain 1 and a type 2/type 3 trinuclear copper cluster at the subunit-subunit interface. The 2dMCO is proposed to be a key intermediate in the evolution of three domain MCOs. The biological function of McoN has not been characterized. Multicopper oxidases couple oxidation of substrates with reduction of dioxygen to water. These MCOs are capable of oxidizing a vast range of substrates, varying from aromatic to inorganic compounds such as metals. |
| cd11024 | CuRO_1_2DMCO_NIR_like | 1.89e-42 | 85 | 206 | 6 | 118 | The cupredoxin domain 1 of a two-domain laccase related to nitrite reductase. The two-domain laccase (small laccase) in this family differs significantly from all laccases. It resembles the two domain nitrite reductase in both sequence and structure. It consists of two cupredoxin domains and forms trimers and hence resembles the quaternary structure of nitrite reductases more than that of large laccases. There are three trinuclear copper clusters in the enzyme localized between domains 1 and 2 of each pair of neighbor chains. Three copper ions of type 1 lie close to one another near the surface of the central part of the trimer, and, effectively, a trimeric substrate binding site is formed in their vicinity. Laccase is a blue multi-copper enzyme that catalyzes the oxidation of a variety of organic substrates coupled to the reduction of molecular oxygen to water. It displays broad substrate specificity, catalyzing the oxidation of a wide variety of aromatic, notably phenolic, and inorganic substances. Laccase has been implicated in a wide spectrum of biological activities. |
| COG2132 | SufI | 8.79e-42 | 69 | 358 | 149 | 448 | Multicopper oxidase with three cupredoxin domains (includes cell division protein FtsP and spore coat protein CotA) [Cell cycle control, cell division, chromosome partitioning, Inorganic ion transport and metabolism, Cell wall/membrane/envelope biogenesis]. |
| COG2132 | SufI | 4.06e-25 | 14 | 359 | 1 | 292 | Multicopper oxidase with three cupredoxin domains (includes cell division protein FtsP and spore coat protein CotA) [Cell cycle control, cell division, chromosome partitioning, Inorganic ion transport and metabolism, Cell wall/membrane/envelope biogenesis]. |
| cd13859 | CuRO_D1_2dMcoN_like | 8.31e-22 | 102 | 205 | 22 | 122 | The first cupredoxin domain of bacterial two domain multicopper oxidase McoN and similar proteins. This family includes bacterial two domain multicopper oxidases (2dMCOs) represented by the McoN from Nitrosomonas europaea. McoN is a trimeric type C blue copper oxidase. Each subunit houses a type 1 copper site in domain 1 and a type 2/type 3 trinuclear copper cluster at the subunit-subunit interface. The 2dMCO is proposed to be a key intermediate in the evolution of three domain MCOs. Its biological function has not been characterized. Multicopper oxidases couple oxidation of substrates with reduction of dioxygen to water. These MCOs are capable of oxidizing a vast range of substrates, varying from aromatic to inorganic compounds such as metals. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QZY04861.1 | 3.86e-224 | 1 | 382 | 1 | 382 |
| QWC19486.1 | 8.22e-179 | 13 | 383 | 13 | 382 |
| QWC20514.1 | 2.90e-169 | 13 | 384 | 13 | 401 |
| CAD7247108.1 | 4.33e-15 | 111 | 365 | 62 | 333 |
| CAE7213185.1 | 5.88e-12 | 115 | 335 | 50 | 266 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3G5W_A | 8.86e-43 | 111 | 367 | 28 | 277 | Crystalstructure of Blue Copper Oxidase from Nitrosomonas europaea [Nitrosomonas europaea],3G5W_B Crystal structure of Blue Copper Oxidase from Nitrosomonas europaea [Nitrosomonas europaea],3G5W_C Crystal structure of Blue Copper Oxidase from Nitrosomonas europaea [Nitrosomonas europaea],3G5W_D Crystal structure of Blue Copper Oxidase from Nitrosomonas europaea [Nitrosomonas europaea],3G5W_E Crystal structure of Blue Copper Oxidase from Nitrosomonas europaea [Nitrosomonas europaea],3G5W_F Crystal structure of Blue Copper Oxidase from Nitrosomonas europaea [Nitrosomonas europaea] |
| 4E9V_A | 3.97e-42 | 111 | 371 | 29 | 282 | MulticopperOxidase mgLAC (data1) [uncultured bacterium],4E9V_B Multicopper Oxidase mgLAC (data1) [uncultured bacterium],4E9V_C Multicopper Oxidase mgLAC (data1) [uncultured bacterium],4E9W_A Multicopper Oxidase mgLAC (data2) [uncultured bacterium],4E9W_B Multicopper Oxidase mgLAC (data2) [uncultured bacterium],4E9W_C Multicopper Oxidase mgLAC (data2) [uncultured bacterium],4E9X_A Multicopper Oxidase mgLAC (data3) [uncultured bacterium],4E9X_B Multicopper Oxidase mgLAC (data3) [uncultured bacterium],4E9X_C Multicopper Oxidase mgLAC (data3) [uncultured bacterium],4E9Y_A Multicopper Oxidase mgLAC (data4) [uncultured bacterium],4E9Y_B Multicopper Oxidase mgLAC (data4) [uncultured bacterium],4E9Y_C Multicopper Oxidase mgLAC (data4) [uncultured bacterium] |
| 3GDC_A | 5.70e-22 | 111 | 344 | 60 | 272 | Crystalstructure of multicopper oxidase [Arthrobacter sp. FB24],3GDC_B Crystal structure of multicopper oxidase [Arthrobacter sp. FB24],3GDC_C Crystal structure of multicopper oxidase [Arthrobacter sp. FB24] |
| 3WIA_A | 1.45e-21 | 101 | 351 | 25 | 266 | Crystalstructure of the N-terminal 1-37 residues deleted mutant of Geobacillus copper nitrite reductase [Geobacillus kaustophilus HTA426],3WIA_B Crystal structure of the N-terminal 1-37 residues deleted mutant of Geobacillus copper nitrite reductase [Geobacillus kaustophilus HTA426],3WIA_C Crystal structure of the N-terminal 1-37 residues deleted mutant of Geobacillus copper nitrite reductase [Geobacillus kaustophilus HTA426],3WIA_D Crystal structure of the N-terminal 1-37 residues deleted mutant of Geobacillus copper nitrite reductase [Geobacillus kaustophilus HTA426],3WIA_E Crystal structure of the N-terminal 1-37 residues deleted mutant of Geobacillus copper nitrite reductase [Geobacillus kaustophilus HTA426],3WIA_F Crystal structure of the N-terminal 1-37 residues deleted mutant of Geobacillus copper nitrite reductase [Geobacillus kaustophilus HTA426],3WIA_G Crystal structure of the N-terminal 1-37 residues deleted mutant of Geobacillus copper nitrite reductase [Geobacillus kaustophilus HTA426],3WIA_H Crystal structure of the N-terminal 1-37 residues deleted mutant of Geobacillus copper nitrite reductase [Geobacillus kaustophilus HTA426],3WIA_I Crystal structure of the N-terminal 1-37 residues deleted mutant of Geobacillus copper nitrite reductase [Geobacillus kaustophilus HTA426] |
| 3WKQ_A | 2.51e-21 | 101 | 351 | 61 | 302 | Copper-containingnitrite reductase from Geobacillus thermodenitrificans in complex with formate [Geobacillus thermodenitrificans NG80-2],3WNI_A 1.50 A resolution crystal structure of dioxygen bound copper-containing nitrite reductase from Geobacillus thermodenitrificans [Geobacillus thermodenitrificans NG80-2],3WNJ_A 1.20 A resolution crystal structure of dioxygen bound copper-containing nitrite reductase from Geobacillus thermodenitrificans [Geobacillus thermodenitrificans NG80-2],4YSA_A Completely oxidized structure of copper nitrite reductase from Geobacillus thermodenitrificans [Geobacillus thermodenitrificans NG80-2],4YSD_A Room temperature structure of copper nitrite reductase from Geobacillus thermodenitrificans [Geobacillus thermodenitrificans NG80-2],4YSO_A Copper nitrite reductase from Geobacillus thermodenitrificans - 0.064 MGy [Geobacillus thermodenitrificans NG80-2],4YSP_A Structure of copper nitrite reductase from Geobacillus thermodenitrificans - 8.32 MGy [Geobacillus thermodenitrificans NG80-2],4YSQ_A Structure of copper nitrite reductase from Geobacillus thermodenitrificans - 8.38 MGy [Geobacillus thermodenitrificans NG80-2],4YSR_A Structure of copper nitrite reductase from Geobacillus thermodenitrificans - 16.6 MGy [Geobacillus thermodenitrificans NG80-2],4YSS_A Structure of copper nitrite reductase from Geobacillus thermodenitrificans - 16.7 MGy [Geobacillus thermodenitrificans NG80-2],4YST_A Structure of copper nitrite reductase from Geobacillus thermodenitrificans - 24.9 MGy [Geobacillus thermodenitrificans NG80-2],4YSU_A Structure of copper nitrite reductase from Geobacillus thermodenitrificans - 25.0 MGy [Geobacillus thermodenitrificans NG80-2],4ZK8_A Copper-containing nitrite reductase from thermophilic bacterium Geobacillus thermodenitrificans (Re-refined) [Geobacillus thermodenitrificans NG80-2],5YTL_A Crystal structure of Geobacillus thermodenitrificans copper-containing nitrite reductase determined with an anaerobically manipulated crystal [Geobacillus thermodenitrificans],6L46_A High-resolution neutron and X-ray joint refined structure of copper-containing nitrite reductase from Geobacillus thermodenitrificans [Geobacillus thermodenitrificans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| I6WZK7 | 7.99e-11 | 13 | 331 | 17 | 325 | Multicopper oxidase MmcO OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=mmcO PE=1 SV=1 |
| Q96UM2 | 1.72e-10 | 115 | 381 | 1 | 266 | Laccase-3 (Fragment) OS=Botryotinia fuckeliana OX=40559 GN=lcc3 PE=3 SV=1 |
| E9F648 | 1.53e-08 | 111 | 240 | 47 | 169 | Laccase 1 OS=Metarhizium robertsii (strain ARSEF 23 / ATCC MYA-3075) OX=655844 GN=Mlac1 PE=2 SV=2 |
| A0A0B4F5S2 | 1.53e-08 | 111 | 240 | 47 | 169 | Laccase 1 OS=Metarhizium brunneum (strain ARSEF 3297) OX=1276141 GN=Mlac1 PE=3 SV=1 |
| A0A0B4F1I0 | 1.53e-08 | 111 | 240 | 47 | 169 | Laccase 1 OS=Metarhizium anisopliae (strain ARSEF 549) OX=1276135 GN=Mlac1 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as TATLIPO
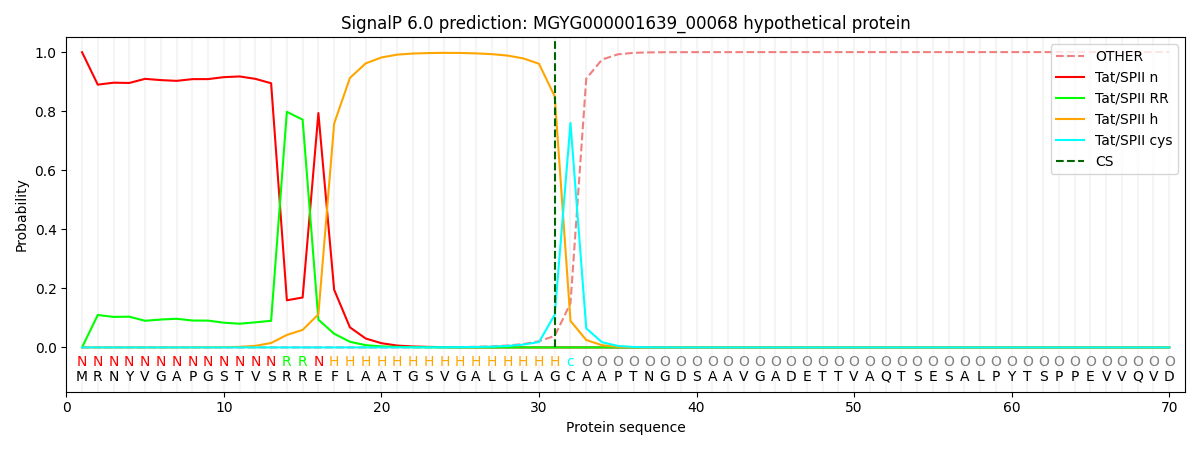
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 0.000038 | 0.000276 | 0.999671 | 0.000000 |
