You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001639_03075
You are here: Home > Sequence: MGYG000001639_03075
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
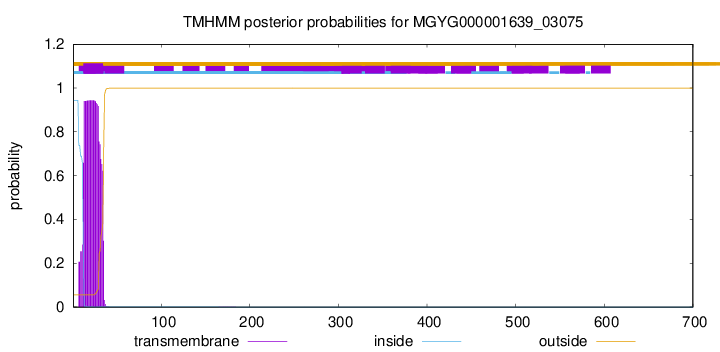
TMHMM annotations
Basic Information help
| Species | Haloferax massiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Archaea; Halobacteriota; Halobacteria; Halobacteriales; Haloferacaceae; Haloferax; Haloferax massiliensis | |||||||||||
| CAZyme ID | MGYG000001639_03075 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 351880; End: 353982 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 375 | 563 | 5.8e-48 | 0.994535519125683 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07081 | DUF1349 | 5.96e-04 | 34 | 188 | 5 | 165 | Protein of unknown function (DUF1349). This family consists of several hypothetical bacterial proteins but contains one sequence from Saccharomyces cerevisiae. Members of this family are typically around 200 residues in length. The function of this family is unknown. |
| cd00063 | FN3 | 0.002 | 221 | 308 | 9 | 93 | Fibronectin type 3 domain; One of three types of internal repeats found in the plasma protein fibronectin. Its tenth fibronectin type III repeat contains an RGD cell recognition sequence in a flexible loop between 2 strands. Approximately 2% of all animal proteins contain the FN3 repeat; including extracellular and intracellular proteins, membrane spanning cytokine receptors, growth hormone receptors, tyrosine phosphatase receptors, and adhesion molecules. FN3-like domains are also found in bacterial glycosyl hydrolases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QOS13727.1 | 0.0 | 1 | 698 | 1 | 697 |
| ADE01341.1 | 0.0 | 1 | 700 | 1 | 699 |
| AKU09480.1 | 0.0 | 37 | 698 | 1 | 661 |
| AEH38979.1 | 8.58e-295 | 1 | 699 | 1 | 693 |
| QLG28824.1 | 1.44e-284 | 1 | 696 | 1 | 700 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as TAT
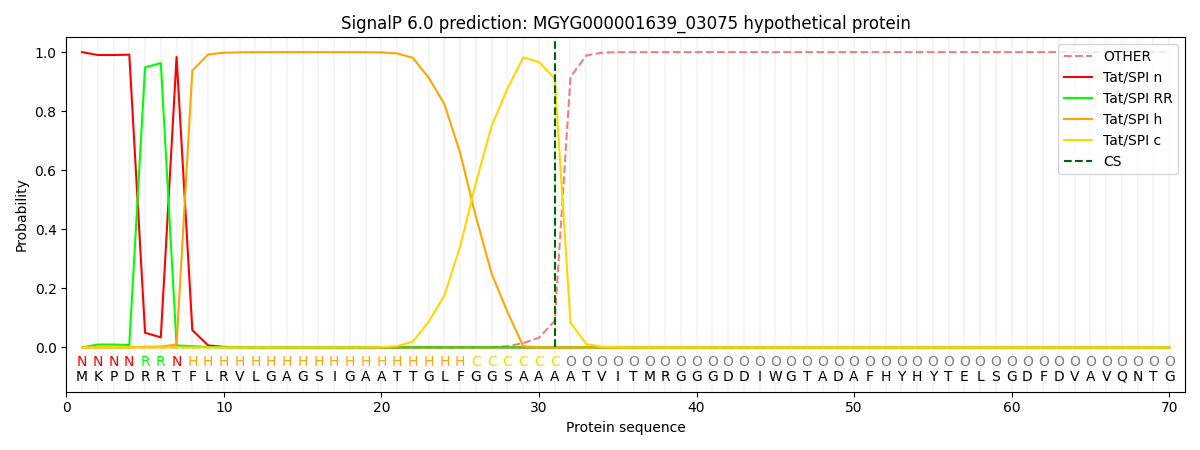
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 0.000000 | 0.999995 | 0.000000 | 0.000000 |