You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001639_03240
You are here: Home > Sequence: MGYG000001639_03240
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Haloferax massiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Archaea; Halobacteriota; Halobacteria; Halobacteriales; Haloferacaceae; Haloferax; Haloferax massiliensis | |||||||||||
| CAZyme ID | MGYG000001639_03240 | |||||||||||
| CAZy Family | CBM16 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 167445; End: 172661 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM16 | 1419 | 1560 | 3.4e-27 | 0.9310344827586207 |
| CBM16 | 1089 | 1228 | 1.3e-25 | 0.896551724137931 |
| CBM16 | 1263 | 1393 | 2.5e-25 | 0.9655172413793104 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam06283 | ThuA | 5.92e-37 | 248 | 436 | 30 | 213 | Trehalose utilisation. This family consists of several bacterial ThuA like proteins. ThuA appears to be involved in utilisation of trehalose. The thuA and thuB genes form part of the trehalose/sucrose transport operon thuEFGKAB, which is located on the pSymB megaplasmid. The thuA and thuB genes are induced in vitro by trehalose but not by sucrose and the extent of its induction depends on the concentration of trehalose available in the medium. |
| TIGR02604 | Piru_Ver_Nterm | 4.17e-24 | 531 | 882 | 21 | 367 | putative membrane-bound dehydrogenase domain. All proteins that score above the trusted cutoff score of 45 to this model are large proteins of either Pirellula sp. 1 or Verrucomicrobium spinosum. These proteins all contain, in addition to this domain, several hundred residues of highly variable sequence, and then a well-conserved C-terminal domain (TIGR02603) that features a putative cytochrome c-type heme binding motif CXXCH. The membrane-bound L-sorbosone dehydrogenase from Acetobacter liquefaciens (Gluconacetobacter liquefaciens) (SP|Q44091) is homologous to this domain but lacks additional sequence regions shared by members of this family and belongs to a different clade of the larger family of homologs. It and its closely related homologs are excluded from the this model by scoring between the trusted (45) and noise (18) cutoffs. |
| COG3828 | COG3828 | 2.23e-11 | 286 | 435 | 65 | 213 | Type 1 glutamine amidotransferase (GATase1)-like domain [General function prediction only]. |
| pfam02018 | CBM_4_9 | 5.26e-08 | 1089 | 1223 | 2 | 114 | Carbohydrate binding domain. This family includes diverse carbohydrate binding domains. |
| pfam02018 | CBM_4_9 | 5.26e-08 | 1262 | 1391 | 2 | 126 | Carbohydrate binding domain. This family includes diverse carbohydrate binding domains. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AKU09421.1 | 0.0 | 1 | 1736 | 1 | 1737 |
| ADE01485.1 | 0.0 | 1 | 1736 | 1 | 1733 |
| QNN20905.1 | 1.18e-246 | 219 | 1255 | 33 | 1001 |
| QOV88701.1 | 2.53e-206 | 215 | 1256 | 33 | 1013 |
| APZ93378.1 | 2.51e-175 | 218 | 1068 | 32 | 874 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| D4GPK6 | 8.82e-06 | 67 | 112 | 533 | 578 | Laccase OS=Haloferax volcanii (strain ATCC 29605 / DSM 3757 / JCM 8879 / NBRC 14742 / NCIMB 2012 / VKM B-1768 / DS2) OX=309800 GN=lccA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as TAT
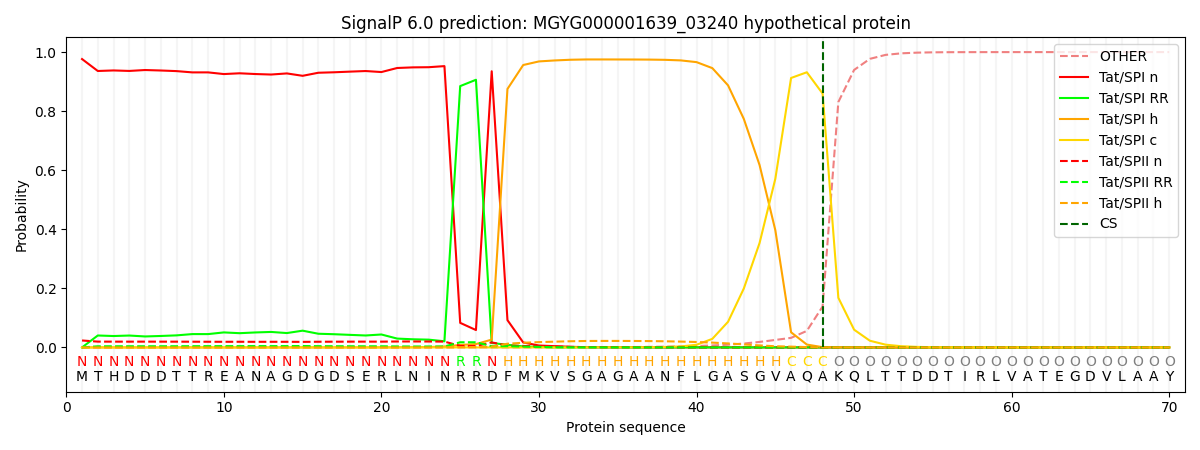
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000006 | 0.000007 | 0.000003 | 0.976457 | 0.023513 | 0.000000 |
