You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001640_00473
You are here: Home > Sequence: MGYG000001640_00473
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Duncaniella sp900549445 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; Duncaniella; Duncaniella sp900549445 | |||||||||||
| CAZyme ID | MGYG000001640_00473 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 20073; End: 22232 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 459 | 714 | 1.6e-47 | 0.6765676567656765 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00633 | Glyco_10 | 1.20e-35 | 460 | 712 | 56 | 263 | Glycosyl hydrolase family 10. |
| pfam00331 | Glyco_hydro_10 | 4.19e-34 | 462 | 714 | 100 | 310 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 6.43e-21 | 460 | 719 | 121 | 344 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam00331 | Glyco_hydro_10 | 6.25e-11 | 61 | 151 | 9 | 98 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 4.79e-05 | 96 | 146 | 1 | 50 | Glycosyl hydrolase family 10. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRQ48288.1 | 6.68e-302 | 11 | 719 | 13 | 719 |
| QUT46089.1 | 6.27e-300 | 11 | 719 | 13 | 719 |
| QIU93949.1 | 2.12e-241 | 1 | 716 | 1 | 725 |
| QDM11106.1 | 2.68e-239 | 1 | 716 | 1 | 725 |
| QUR44907.1 | 3.93e-239 | 1 | 716 | 1 | 725 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4MGS_A | 2.86e-36 | 154 | 299 | 1 | 150 | BiXyn10ACBM1 APO [Bacteroides intestinalis DSM 17393] |
| 4QPW_A | 2.72e-32 | 164 | 296 | 5 | 141 | BiXyn10ACBM1 with Xylohexaose Bound [Bacteroides intestinalis DSM 17393] |
| 4W8L_A | 1.90e-16 | 452 | 714 | 100 | 343 | Structureof GH10 from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4W8L_B Structure of GH10 from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4W8L_C Structure of GH10 from Paenibacillus barcinonensis [Paenibacillus barcinonensis] |
| 6FHE_A | 2.21e-16 | 450 | 709 | 114 | 335 | Highlyactive enzymes by automated modular backbone assembly and sequence design [synthetic construct] |
| 2F8Q_A | 1.15e-14 | 459 | 632 | 118 | 263 | Analkali thermostable F/10 xylanase from alkalophilic Bacillus sp. NG-27 [Bacillus sp. NG-27],2F8Q_B An alkali thermostable F/10 xylanase from alkalophilic Bacillus sp. NG-27 [Bacillus sp. NG-27] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q4P902 | 3.09e-15 | 462 | 709 | 139 | 332 | Endo-1,4-beta-xylanase UM03411 OS=Ustilago maydis (strain 521 / FGSC 9021) OX=237631 GN=UMAG_03411 PE=1 SV=1 |
| O69230 | 6.03e-15 | 452 | 714 | 466 | 709 | Endo-1,4-beta-xylanase C OS=Paenibacillus barcinonensis OX=198119 GN=xynC PE=1 SV=1 |
| Q9HEZ1 | 7.22e-14 | 462 | 715 | 195 | 405 | Endo-1,4-beta-xylanase A OS=Phanerodontia chrysosporium OX=2822231 GN=xynA PE=1 SV=1 |
| Q59675 | 8.27e-14 | 478 | 718 | 374 | 599 | Endo-beta-1,4-xylanase Xyn10C OS=Cellvibrio japonicus OX=155077 GN=xyn10C PE=1 SV=2 |
| P40942 | 2.63e-13 | 450 | 714 | 146 | 381 | Thermostable celloxylanase OS=Thermoclostridium stercorarium OX=1510 GN=xynB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
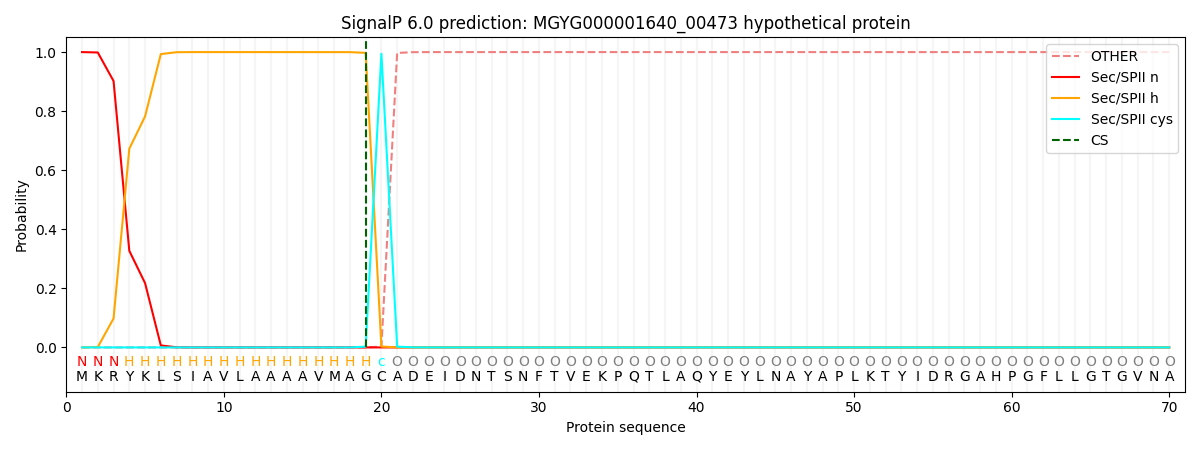
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000039 | 0.000000 | 0.000000 | 0.000000 |
