You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001641_00395
You are here: Home > Sequence: MGYG000001641_00395
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-485 sp900760815 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-485; CAG-485 sp900760815 | |||||||||||
| CAZyme ID | MGYG000001641_00395 | |||||||||||
| CAZy Family | GH136 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 94664; End: 96196 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH136 | 23 | 442 | 8.5e-92 | 0.8329938900203666 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13229 | Beta_helix | 2.35e-07 | 149 | 323 | 8 | 144 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam13229 | Beta_helix | 7.29e-04 | 221 | 389 | 8 | 139 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ALJ60891.1 | 2.39e-165 | 1 | 506 | 4 | 506 |
| QEW35088.1 | 6.75e-143 | 15 | 503 | 14 | 499 |
| ALK82684.1 | 6.75e-143 | 15 | 503 | 14 | 499 |
| QUT56379.1 | 6.75e-143 | 15 | 503 | 14 | 499 |
| AND18533.1 | 4.77e-142 | 15 | 503 | 10 | 495 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6KQT_A | 1.94e-30 | 26 | 352 | 247 | 617 | CrystalStructure of GH136 lacto-N-biosidase from Eubacterium ramulus - native protein [Eubacterium ramulus ATCC 29099] |
| 6KQS_A | 2.61e-30 | 26 | 352 | 247 | 617 | CrystalStructure of GH136 lacto-N-biosidase from Eubacterium ramulus - selenomethionine derivative [Eubacterium ramulus ATCC 29099] |
| 7V6I_A | 1.20e-19 | 24 | 423 | 14 | 521 | ChainA, Lacto-N-biosidase [Bifidobacterium saguini DSM 23967] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
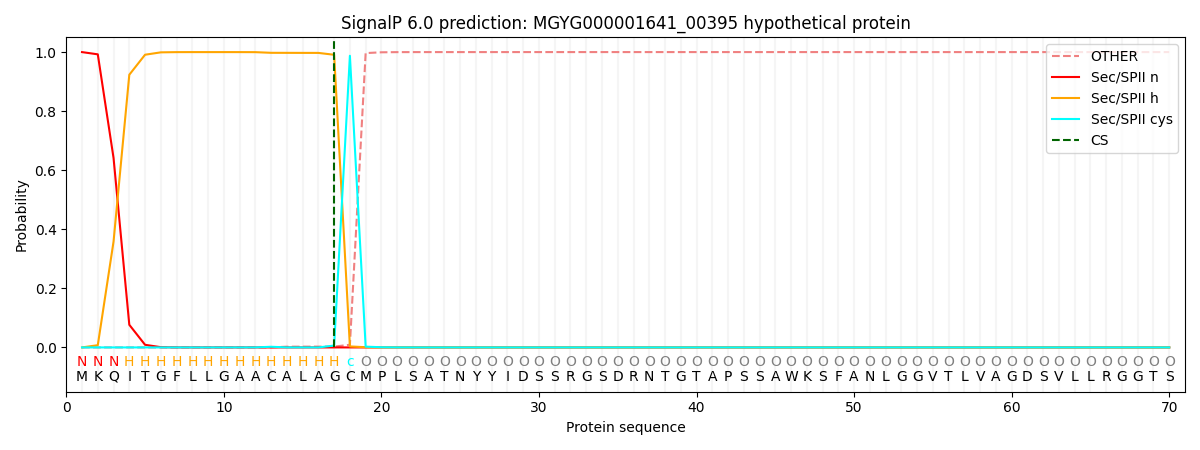
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000041 | 0.000000 | 0.000000 | 0.000000 |
