You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001643_01546
You are here: Home > Sequence: MGYG000001643_01546
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
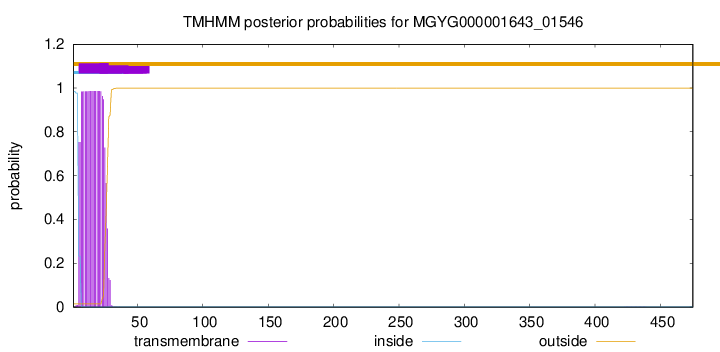
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-485; | |||||||||||
| CAZyme ID | MGYG000001643_01546 | |||||||||||
| CAZy Family | GH39 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 15296; End: 16723 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH39 | 85 | 245 | 1.4e-30 | 0.3874709976798144 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01229 | Glyco_hydro_39 | 1.83e-11 | 92 | 262 | 74 | 263 | Glycosyl hydrolases family 39. |
| pfam02449 | Glyco_hydro_42 | 3.72e-08 | 73 | 185 | 16 | 169 | Beta-galactosidase. This group of beta-galactosidase enzymes belong to the glycosyl hydrolase 42 family. The enzyme catalyzes the hydrolysis of terminal, non-reducing terminal beta-D-galactosidase residues. |
| pfam00331 | Glyco_hydro_10 | 8.29e-06 | 82 | 200 | 46 | 164 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 3.32e-05 | 82 | 200 | 69 | 188 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDO70685.1 | 4.19e-205 | 29 | 467 | 38 | 475 |
| AEI51085.1 | 2.49e-132 | 33 | 467 | 42 | 493 |
| AEI51083.1 | 1.06e-130 | 32 | 467 | 45 | 478 |
| AQG80712.1 | 1.09e-129 | 49 | 467 | 76 | 510 |
| CCH00422.1 | 2.18e-126 | 32 | 467 | 48 | 486 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4EKJ_A | 1.58e-08 | 46 | 239 | 27 | 231 | ChainA, Beta-xylosidase [Caulobacter vibrioides] |
| 4M29_A | 1.58e-08 | 46 | 239 | 27 | 231 | Structureof a GH39 Beta-xylosidase from Caulobacter crescentus [Caulobacter vibrioides CB15] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
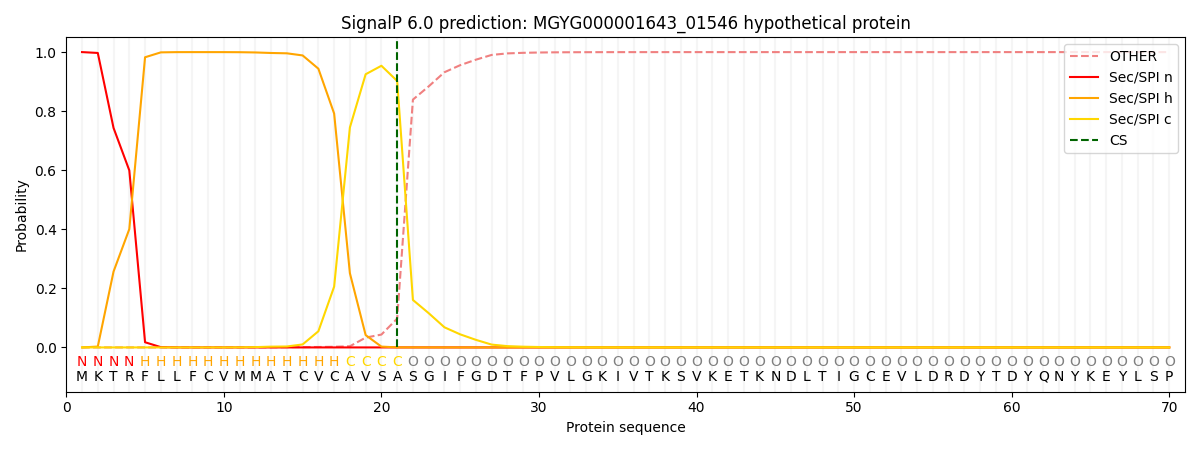
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000287 | 0.999104 | 0.000171 | 0.000146 | 0.000130 | 0.000127 |