You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001649_02016
Basic Information
help
| Species |
UBA1409 sp900760125
|
| Lineage |
Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; UBA1409; UBA1409 sp900760125
|
| CAZyme ID |
MGYG000001649_02016
|
| CAZy Family |
GH43 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 657 |
|
74191.46 |
4.799 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000001649 |
2605663 |
MAG |
United States |
North America |
|
| Gene Location |
Start: 137;
End: 2110
Strand: -
|
No EC number prediction in MGYG000001649_02016.
MGYG000001649_02016 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| QTE67541.1
|
1.94e-188 |
46 |
653 |
39 |
648 |
| AHF25595.1
|
4.57e-181 |
48 |
653 |
47 |
654 |
This protein is predicted as OTHER
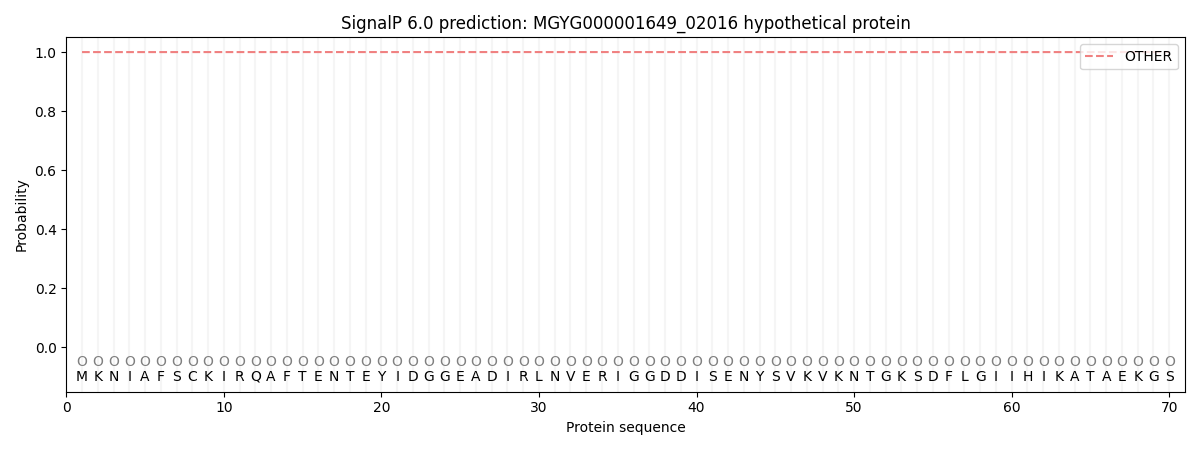
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
1.000085
|
0.000000
|
0.000000
|
0.000000
|
0.000000
|
0.000000
|
There is no transmembrane helices in MGYG000001649_02016.

