You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001652_01673
You are here: Home > Sequence: MGYG000001652_01673
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | V9D3004 sp002349525 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; V9D3004; V9D3004 sp002349525 | |||||||||||
| CAZyme ID | MGYG000001652_01673 | |||||||||||
| CAZy Family | CE1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 231; End: 1574 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE1 | 205 | 443 | 1.7e-46 | 0.9691629955947136 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG2382 | Fes | 7.85e-30 | 152 | 447 | 23 | 297 | Enterochelin esterase or related enzyme [Inorganic ion transport and metabolism]. |
| pfam00756 | Esterase | 7.98e-24 | 205 | 438 | 1 | 239 | Putative esterase. This family contains Esterase D. However it is not clear if all members of the family have the same function. This family is related to the pfam00135 family. |
| COG2819 | YbbA | 9.99e-14 | 208 | 357 | 19 | 172 | Predicted hydrolase of the alpha/beta superfamily [General function prediction only]. |
| COG0627 | FrmB | 1.63e-12 | 205 | 435 | 28 | 298 | S-formylglutathione hydrolase FrmB [Defense mechanisms]. |
| PLN02442 | PLN02442 | 1.07e-09 | 196 | 429 | 16 | 261 | S-formylglutathione hydrolase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADL52399.1 | 9.24e-85 | 176 | 447 | 58 | 323 |
| ADZ84563.1 | 1.03e-71 | 184 | 447 | 216 | 473 |
| QEH70018.1 | 2.85e-71 | 184 | 447 | 216 | 473 |
| QIA08067.1 | 7.34e-52 | 181 | 447 | 26 | 278 |
| QEH42336.1 | 1.81e-51 | 174 | 447 | 17 | 276 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5CXU_A | 9.55e-53 | 183 | 446 | 11 | 270 | Structureof the CE1 ferulic acid esterase AmCE1/Fae1A, from the anaerobic fungi Anaeromyces mucronatus in the absence of substrate [Anaeromyces mucronatus],5CXX_A Structure of a CE1 ferulic acid esterase, AmCE1/Fae1A, from Anaeromyces mucronatus in complex with Ferulic acid [Anaeromyces mucronatus],5CXX_B Structure of a CE1 ferulic acid esterase, AmCE1/Fae1A, from Anaeromyces mucronatus in complex with Ferulic acid [Anaeromyces mucronatus],5CXX_C Structure of a CE1 ferulic acid esterase, AmCE1/Fae1A, from Anaeromyces mucronatus in complex with Ferulic acid [Anaeromyces mucronatus] |
| 1JJF_A | 1.19e-45 | 180 | 444 | 14 | 256 | ChainA, Endo-1,4-beta-xylanase Z [Acetivibrio thermocellus] |
| 1JT2_A | 3.26e-45 | 180 | 444 | 14 | 256 | ChainA, PROTEIN (ENDO-1,4-BETA-XYLANASE Z) [Acetivibrio thermocellus] |
| 7B5V_A | 7.27e-30 | 184 | 447 | 117 | 386 | ChainA, Carbohydrate Esterase family 1 protein with an N-terminal carbohydrate binding module family 48 [Dysgonomonas mossii DSM 22836],7B5V_B Chain B, Carbohydrate Esterase family 1 protein with an N-terminal carbohydrate binding module family 48 [Dysgonomonas mossii DSM 22836],7B6B_A Chain A, Carbohydrate Esterase family 1 protein with an N-terminal carbohydrate binding module family 48 [Dysgonomonas mossii DSM 22836],7B6B_B Chain B, Carbohydrate Esterase family 1 protein with an N-terminal carbohydrate binding module family 48 [Dysgonomonas mossii DSM 22836] |
| 6MOT_A | 1.66e-23 | 184 | 447 | 95 | 359 | ChainA, Isoamylase N-terminal domain protein [Bacteroides intestinalis DSM 17393] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P10478 | 2.00e-41 | 180 | 444 | 33 | 275 | Endo-1,4-beta-xylanase Z OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=xynZ PE=1 SV=3 |
| D5EXZ4 | 2.36e-20 | 190 | 447 | 423 | 670 | Carbohydrate acetyl esterase/feruloyl esterase OS=Prevotella ruminicola (strain ATCC 19189 / JCM 8958 / 23) OX=264731 GN=axe1-6A PE=1 SV=1 |
| D5EY13 | 9.53e-13 | 186 | 382 | 488 | 666 | Endo-1,4-beta-xylanase/feruloyl esterase OS=Prevotella ruminicola (strain ATCC 19189 / JCM 8958 / 23) OX=264731 GN=xyn10D-fae1A PE=1 SV=1 |
| P9WM38 | 9.90e-09 | 202 | 337 | 178 | 301 | Esterase MT1326 OS=Mycobacterium tuberculosis (strain CDC 1551 / Oshkosh) OX=83331 GN=MT1326 PE=3 SV=1 |
| P9WM39 | 9.90e-09 | 202 | 337 | 178 | 301 | Esterase Rv1288 OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=Rv1288 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
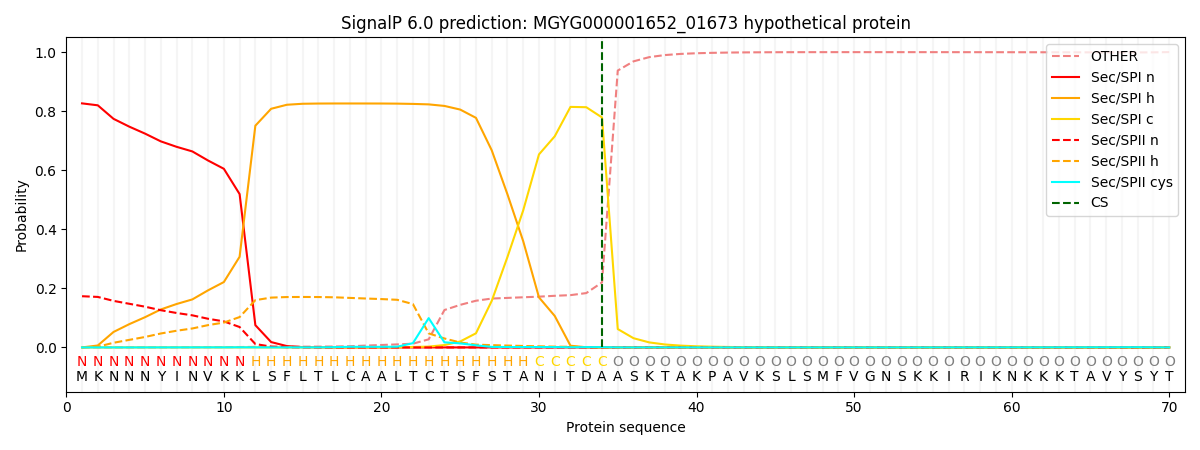
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000784 | 0.819816 | 0.178554 | 0.000327 | 0.000265 | 0.000212 |
