You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001655_00352
You are here: Home > Sequence: MGYG000001655_00352
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Butyricimonas faecalis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Marinifilaceae; Butyricimonas; Butyricimonas faecalis | |||||||||||
| CAZyme ID | MGYG000001655_00352 | |||||||||||
| CAZy Family | GT83 | |||||||||||
| CAZyme Description | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 95974; End: 97569 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT83 | 8 | 428 | 1.1e-67 | 0.7981481481481482 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1807 | ArnT | 7.37e-25 | 7 | 410 | 15 | 422 | 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| PRK13279 | arnT | 4.73e-12 | 1 | 413 | 2 | 429 | lipid IV(A) 4-amino-4-deoxy-L-arabinosyltransferase. |
| pfam13231 | PMT_2 | 8.65e-08 | 55 | 203 | 1 | 145 | Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRO50573.1 | 0.0 | 1 | 512 | 1 | 512 |
| QKH86812.1 | 2.31e-144 | 7 | 507 | 8 | 512 |
| QLK84536.1 | 2.31e-144 | 7 | 507 | 8 | 512 |
| QUU03965.1 | 3.27e-144 | 7 | 507 | 8 | 512 |
| QTO25703.1 | 2.63e-143 | 7 | 507 | 8 | 512 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O67270 | 2.00e-14 | 10 | 316 | 6 | 311 | Uncharacterized protein aq_1220 OS=Aquifex aeolicus (strain VF5) OX=224324 GN=aq_1220 PE=3 SV=1 |
| Q1CIH9 | 1.85e-10 | 7 | 313 | 15 | 321 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Yersinia pestis bv. Antiqua (strain Nepal516) OX=377628 GN=arnT PE=3 SV=1 |
| Q1C744 | 1.85e-10 | 7 | 313 | 15 | 321 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Yersinia pestis bv. Antiqua (strain Antiqua) OX=360102 GN=arnT PE=3 SV=1 |
| Q8ZDX9 | 1.85e-10 | 7 | 313 | 15 | 321 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Yersinia pestis OX=632 GN=arnT PE=3 SV=1 |
| Q66A06 | 1.85e-10 | 7 | 313 | 15 | 321 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Yersinia pseudotuberculosis serotype I (strain IP32953) OX=273123 GN=arnT PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.985698 | 0.013907 | 0.000218 | 0.000048 | 0.000042 | 0.000086 |
TMHMM Annotations download full data without filtering help
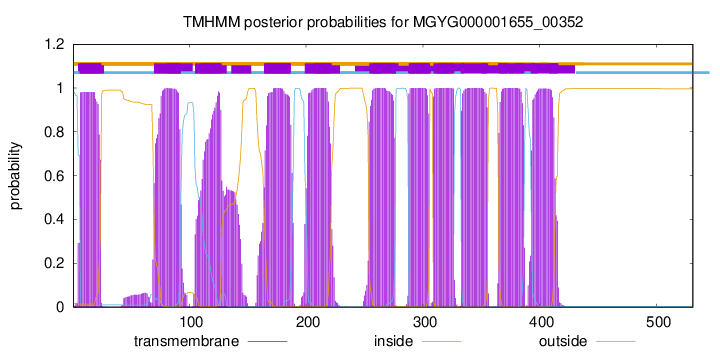
| start | end |
|---|---|
| 7 | 24 |
| 70 | 92 |
| 105 | 127 |
| 164 | 186 |
| 199 | 221 |
| 254 | 276 |
| 288 | 305 |
| 309 | 326 |
| 333 | 355 |
| 365 | 386 |
| 393 | 415 |
