You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001657_00300
You are here: Home > Sequence: MGYG000001657_00300
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | RC9 sp900760425 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; UBA932; RC9; RC9 sp900760425 | |||||||||||
| CAZyme ID | MGYG000001657_00300 | |||||||||||
| CAZy Family | GH78 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 18609; End: 22049 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH78 | 633 | 835 | 6e-16 | 0.376984126984127 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3408 | GDB1 | 1.32e-36 | 544 | 980 | 221 | 641 | Glycogen debranching enzyme (alpha-1,6-glucosidase) [Carbohydrate transport and metabolism]. |
| pfam04685 | DUF608 | 7.50e-09 | 677 | 775 | 99 | 195 | Glycosyl-hydrolase family 116, catalytic region. This represents a family of archaeal, bacterial and eukaryotic glycosyl hydrolases, that belong to superfamily GH116. The primary catabolic pathway for glucosylceramide is catalysis by the lysosomal enzyme glucocerebrosidase. In higher eukaryotes, glucosylceramide is the precursor of glycosphingolipids, a complex group of ubiquitous membrane lipids. Mutations in the human protein cause motor-neurone defects in hereditary spastic paraplegia. The catalytic nucleophile, identified in UniProtKB:Q97YG8_SULSO, is a glutamine-335, with the likely acid/base at Asp-442 and the aspartates at Asp-406 and Asp-458 residues also playing a role in the catalysis of glucosides and xylosides that are beta-bound to hydrophobic groups. The family is defined as GH116, which presently includes enzymes with beta-glucosidase, EC:3.2.1.21, beta-xylosidase, EC:3.2.1.37, and glucocerebrosidase EC:3.2.1.45 activity. |
| pfam06202 | GDE_C | 2.00e-08 | 673 | 928 | 80 | 368 | Amylo-alpha-1,6-glucosidase. This family includes human glycogen branching enzyme AGL. This enzyme contains a number of distinct catalytic activities. It has been shown for the yeast homolog GDB1 that mutations in this region disrupt the enzymes Amylo-alpha-1,6-glucosidase (EC:3.2.1.33). |
| COG3387 | SGA1 | 2.00e-07 | 620 | 928 | 284 | 593 | Glucoamylase (glucan-1,4-alpha-glucosidase), GH15 family [Carbohydrate transport and metabolism]. |
| COG4354 | COG4354 | 1.29e-05 | 677 | 889 | 442 | 668 | Uncharacterized protein, contains GBA2_N and DUF608 domains [Function unknown]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| APF19554.1 | 3.62e-184 | 86 | 1129 | 74 | 1107 |
| BBM70086.1 | 2.94e-97 | 308 | 1133 | 14 | 836 |
| ACY48648.1 | 6.05e-97 | 308 | 1120 | 33 | 843 |
| AEN73635.1 | 1.15e-96 | 308 | 1133 | 33 | 855 |
| AHG91625.1 | 8.22e-69 | 326 | 1106 | 58 | 838 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
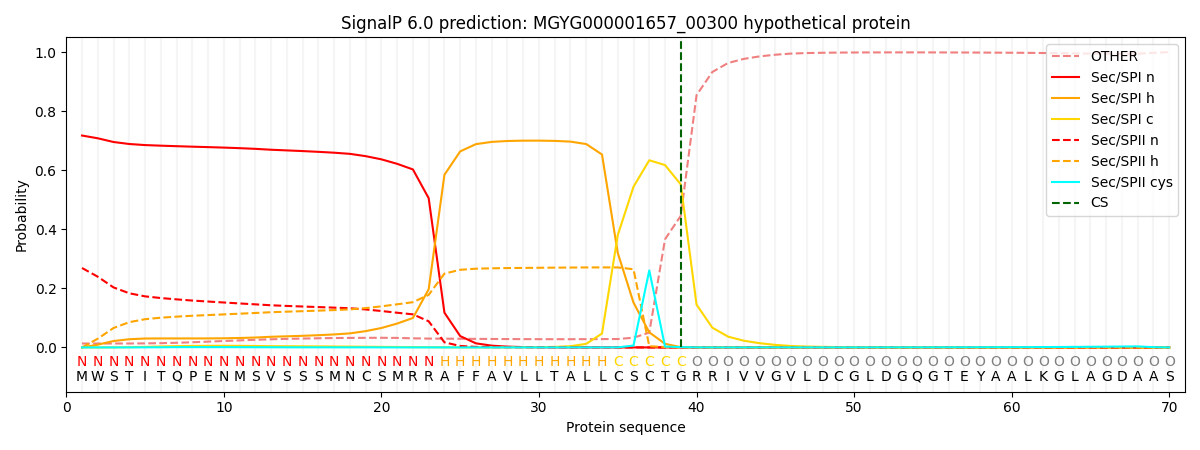
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.015960 | 0.687389 | 0.294539 | 0.000590 | 0.000923 | 0.000584 |
