You are browsing environment: HUMAN GUT
MGYG000001657_00800
Basic Information
help
Species
RC9 sp900760425
Lineage
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; UBA932; RC9; RC9 sp900760425
CAZyme ID
MGYG000001657_00800
CAZy Family
GH49
CAZyme Description
hypothetical protein
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
531
59201.1
4.8609
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000001657
2980281
MAG
United States
North America
Gene Location
Start: 6830;
End: 8425
Strand: +
No EC number prediction in MGYG000001657_00800.
Family
Start
End
Evalue
family coverage
GH49
178
516
2.5e-49
0.6484517304189436
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam17433
Glyco_hydro_49N
6.31e-05
108
187
81
185
Glycosyl hydrolase family 49 N-terminal Ig-like domain. Family of dextranase (EC 3.2.1.11) and isopullulanase (EC 3.2.1.57). Dextranase hydrolyzes alpha-1,6-glycosidic bonds in dextran polymers. This domain corresponds to the N-terminal Ig-like fold.
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
6NZS_A
4.95e-34
98
530
72
574
DextranaseAoDex KQ11 [Pseudarthrobacter oxydans]
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
P70744
4.22e-35
56
530
79
625
Dextranase OS=Arthrobacter globiformis OX=1665 PE=3 SV=1
more
P39652
1.76e-32
56
530
79
625
Dextranase OS=Arthrobacter sp. (strain CB-8) OX=74565 PE=1 SV=1
more
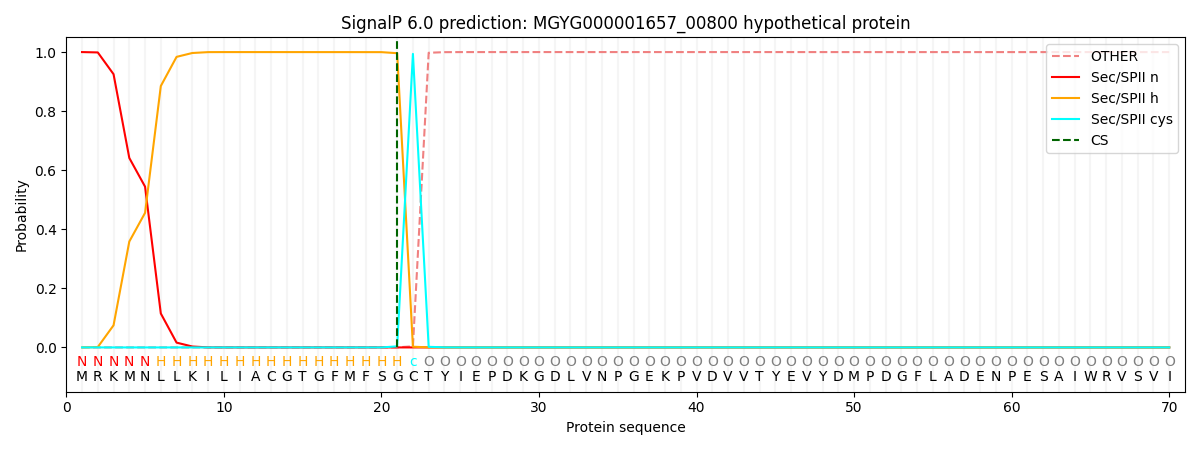
This protein is predicted as LIPO
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.000000
0.000000
1.000069
0.000000
0.000000
0.000000
There is no transmembrane helices in MGYG000001657_00800.