You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001659_00819
You are here: Home > Sequence: MGYG000001659_00819
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paraprevotella sp900760455 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Paraprevotella; Paraprevotella sp900760455 | |||||||||||
| CAZyme ID | MGYG000001659_00819 | |||||||||||
| CAZy Family | CBM13 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 29327; End: 32128 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM13 | 399 | 532 | 1.3e-17 | 0.6914893617021277 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13385 | Laminin_G_3 | 2.07e-35 | 678 | 834 | 1 | 151 | Concanavalin A-like lectin/glucanases superfamily. This domain belongs to the Concanavalin A-like lectin/glucanases superfamily. |
| pfam14200 | RicinB_lectin_2 | 1.92e-14 | 434 | 521 | 4 | 85 | Ricin-type beta-trefoil lectin domain-like. |
| pfam14200 | RicinB_lectin_2 | 2.31e-09 | 484 | 535 | 1 | 58 | Ricin-type beta-trefoil lectin domain-like. |
| pfam00652 | Ricin_B_lectin | 4.72e-08 | 452 | 537 | 4 | 90 | Ricin-type beta-trefoil lectin domain. |
| cd00161 | RICIN | 1.05e-07 | 402 | 531 | 1 | 124 | Ricin-type beta-trefoil; Carbohydrate-binding domain formed from presumed gene triplication. The domain is found in a variety of molecules serving diverse functions such as enzymatic activity, inhibitory toxicity and signal transduction. Highly specific ligand binding occurs on exposed surfaces of the compact domain sturcture. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT88791.1 | 0.0 | 2 | 933 | 12 | 940 |
| ALJ60208.1 | 0.0 | 2 | 933 | 12 | 940 |
| ASB47762.1 | 2.11e-294 | 25 | 932 | 31 | 932 |
| APZ45922.1 | 2.14e-238 | 7 | 933 | 12 | 930 |
| AUC19784.1 | 2.14e-238 | 7 | 933 | 12 | 930 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5HON_A | 1.14e-12 | 637 | 841 | 5 | 209 | Structureof Domain 4 of AbnA, a GH43 extracellular arabinanase from Geobacillus stearothermophilus, in complex with arabinotriose [Geobacillus stearothermophilus],5HON_B Structure of Domain 4 of AbnA, a GH43 extracellular arabinanase from Geobacillus stearothermophilus, in complex with arabinotriose [Geobacillus stearothermophilus] |
| 5HO0_A | 2.17e-11 | 637 | 841 | 644 | 848 | Crystalstructure of AbnA (closed conformation), a GH43 extracellular arabinanase from Geobacillus stearothermophilus [Geobacillus stearothermophilus],5HO2_A Crystal structure of AbnA (open conformation), a GH43 extracellular arabinanase from Geobacillus stearothermophilus [Geobacillus stearothermophilus],5HOF_A Crystal structure of AbnA, a GH43 extracellular arabinanase from Geobacillus stearothermophilus, in complex with arabinopentaose [Geobacillus stearothermophilus],5HP6_A Structure of AbnA, a GH43 extracellular arabinanase from Geobacillus stearothermophilus (a new conformational state) [Geobacillus stearothermophilus] |
| 5G56_A | 6.51e-11 | 366 | 620 | 434 | 695 | ChainA, Carbohydrate Binding Family 6 [Acetivibrio thermocellus] |
| 3MPC_A | 3.31e-08 | 540 | 620 | 2 | 83 | ChainA, Fn3-like protein [Acetivibrio thermocellus ATCC 27405],3MPC_B Chain B, Fn3-like protein [Acetivibrio thermocellus ATCC 27405] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P16950 | 8.48e-07 | 536 | 629 | 922 | 1014 | Amylopullulanase OS=Thermoanaerobacter thermohydrosulfuricus OX=1516 GN=apu PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
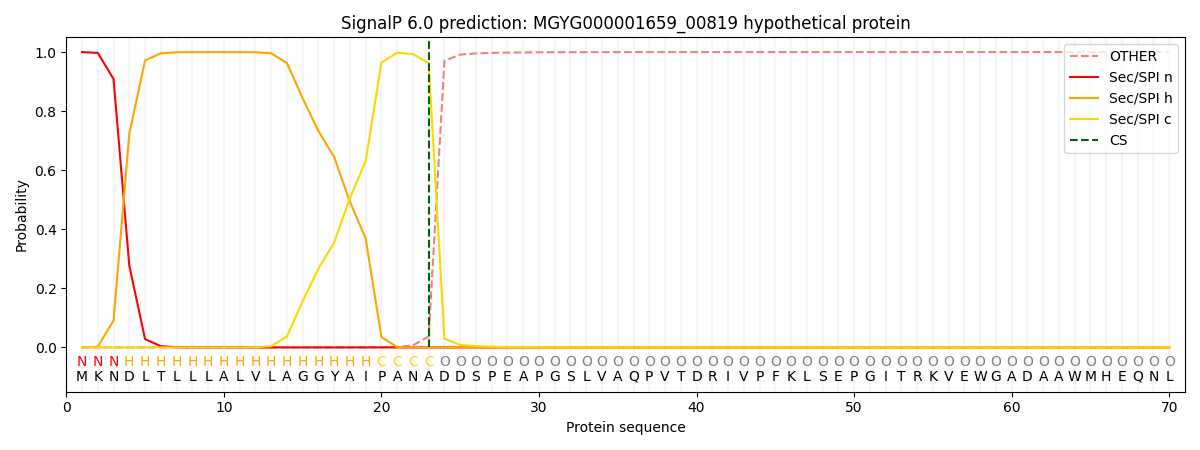
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000400 | 0.998684 | 0.000237 | 0.000265 | 0.000211 | 0.000177 |
