You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001659_02594
You are here: Home > Sequence: MGYG000001659_02594
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paraprevotella sp900760455 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Paraprevotella; Paraprevotella sp900760455 | |||||||||||
| CAZyme ID | MGYG000001659_02594 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1750; End: 3183 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH28 | 191 | 455 | 1.3e-39 | 0.7876923076923077 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5434 | Pgu1 | 8.99e-08 | 132 | 452 | 83 | 418 | Polygalacturonase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIU95271.1 | 1.74e-191 | 15 | 475 | 7 | 467 |
| QUT22798.1 | 3.68e-186 | 19 | 475 | 12 | 468 |
| QRQ56356.1 | 3.68e-186 | 19 | 475 | 12 | 468 |
| ALJ44674.1 | 3.68e-186 | 19 | 475 | 12 | 468 |
| SCV09008.1 | 3.68e-186 | 19 | 475 | 12 | 468 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1WMR_A | 2.58e-06 | 24 | 351 | 37 | 403 | CrystalStructure of Isopullulanase from Aspergillus niger ATCC 9642 [Aspergillus niger],1WMR_B Crystal Structure of Isopullulanase from Aspergillus niger ATCC 9642 [Aspergillus niger],1X0C_A Improved Crystal Structure of Isopullulanase from Aspergillus niger ATCC 9642 [Aspergillus niger],1X0C_B Improved Crystal Structure of Isopullulanase from Aspergillus niger ATCC 9642 [Aspergillus niger],2Z8G_A Aspergillus niger ATCC9642 isopullulanase complexed with isopanose [Aspergillus niger],2Z8G_B Aspergillus niger ATCC9642 isopullulanase complexed with isopanose [Aspergillus niger] |
| 3WWG_A | 2.58e-06 | 24 | 351 | 37 | 403 | Crystalstructure of the N-glycan-deficient variant N448A of isopullulanase complexed with isopanose [Aspergillus niger],3WWG_B Crystal structure of the N-glycan-deficient variant N448A of isopullulanase complexed with isopanose [Aspergillus niger],3WWG_C Crystal structure of the N-glycan-deficient variant N448A of isopullulanase complexed with isopanose [Aspergillus niger],3WWG_D Crystal structure of the N-glycan-deficient variant N448A of isopullulanase complexed with isopanose [Aspergillus niger] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
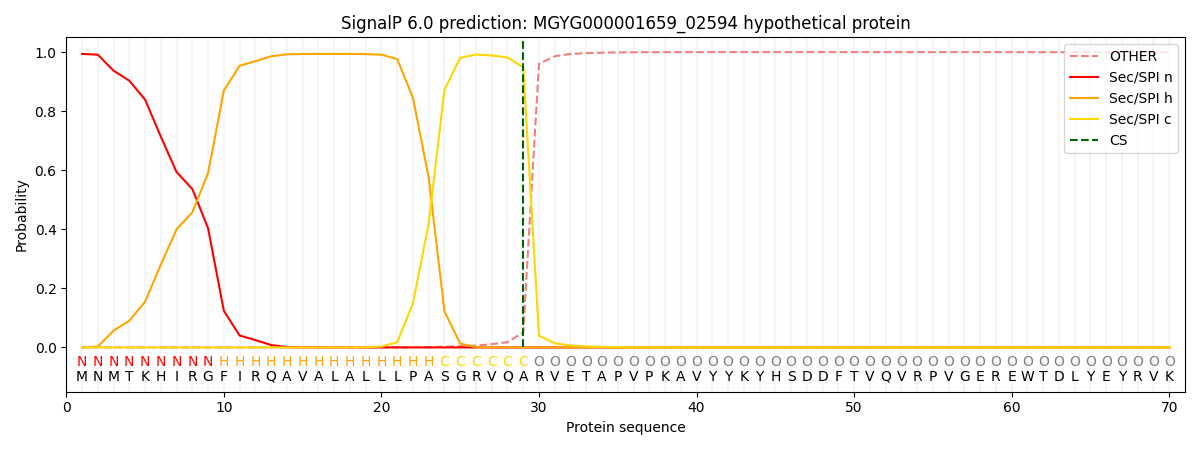
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001847 | 0.989401 | 0.007565 | 0.000600 | 0.000335 | 0.000234 |
